As the first agricultural animal to complete whole genome sequencing, the chicken is an important economic animal and an important model organism in basic biology and molecular genetics research. The mechanism of ovarian cancer in chickens is similar to that of human ovarian cancer, so chickens can be used as an essential model for research on the pathogenesis of ovarian reproductive diseases such as human ovarian cancer.
Chicken embryos have been used as model organisms to study the developing embryo because of their ease of access, manipulation, culture, and observation. This has important implications for the study of organ formation and cell differentiation. In addition to evolutionary development, chicken embryos have been used to develop drugs for anti-cancer and cardiovascular diseases. Even chicken embryos have been used to produce special medical proteins. With the broadening of scientific research, ChIP-seq technology is increasingly being applied to chicken-related studies.
Lifeasible is a highly regarded biotechnology company with a standard animal-oriented sequencing system, and our team of scientists can provide you with professional chicken ChIP-seq services. Your satisfaction is our aim, and we focus on providing you with solutions to uncover the DNA sites that bind specifically to target proteins in chicken.

Although ChIP-seq has less available information on chicken, with the advancement of molecular biology, ChIP-seq explores various biological phenomena with higher resolution. Based on our rich sequencing experience, we have summarized a complete ChIP-seq process for application with chicken. And we have unique processing methods for quality control, comparison, peak calling, and super-enhancer identification of sequencing data.
We have developed a workflow for ChIP-seq analysis of chicken skin appendages to provide technical support for chicken research, especially for systems biology analysis. Data quality is one of the most critical issues in ChIP-seq analysis, and our service process completely overcomes this challenge.
We performed high-throughput sequencing of H3K4me3 and H3K27me3 in chicken follicular mesenchymal cells and male germ cells using ChIP-seq technology to investigate the epigenetic control of gene expression in these two cells. The final analysis of the obtained ChIP-seq data demonstrated that the bivalent state of H3K4me3/H3K27me3 has a repressive effect on germ cell transcription.
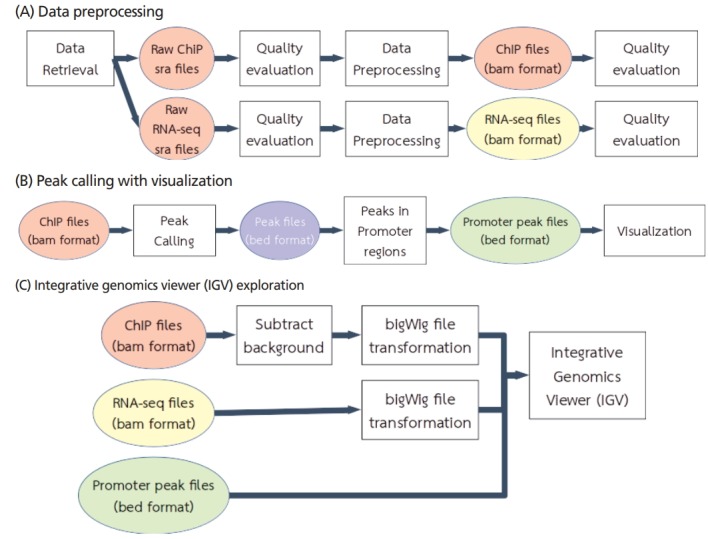 Figure 1. Workflow of ChIP-seq data acquisition and analysis. (Chokeshaiusaha, K, et al. 2018)
Figure 1. Workflow of ChIP-seq data acquisition and analysis. (Chokeshaiusaha, K, et al. 2018)
To verify the role of Spin1z in chicken male differentiation, we performed ChIP-seq on male embryonic stem cells (ESCs) and primordial germ cells (PGCs). Data analysis showed that Spin1z promotes chicken male differentiation by repressing Tcf4 transcription.
It is recommended to provide two sample preparations, if possible, to ensure the quality and continuity of the experiment.
Lifeasible's animal-oriented established sequencing technology platform can provide you with a comprehensive chicken ChIP-seq solution to help you start your research on chicken systems biology, reproduction, and other aspects to enhance the role of chickens in evolutionary developmental processes, medicine, and drug development. Meeting all your needs is always our service philosophy. Please feel free to contact us for questions, inquiries, or collaboration.
References
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
Why Do Plants Blush When They Are Hungry?
April 26, 2024
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024