With the rapid development of plant genome editing technology, especially the application of CRISPR-Cas system, precise plant genetic improvement has become possible. Although the CRISPR-Cas system shows great potential in plant genome editing, its application still faces some challenges, especially the differences in editing efficiency of different guide RNAs (gRNAs), which increases the difficulty of screening editing events. Traditional strategies often rely on marker genes (such as antibiotic or herbicide resistance markers) to screen for transgenic events. However, these marker genes do not directly reflect the editing status of the target gene, and researchers need to further screen the transformation events to determine which events actually occur for the desired editing. Previous studies have used surrogate reporter systems to improve genome editing efficiency. However, because different sgRNAs are used to edit surrogate reporter genes and endogenous genes respectively, these systems can only screen events with high CRISPR-Cas expression, but fail to effectively display the editing activity of endogenous gene sites. Therefore, there is an urgent need to develop more integrated and effective methods to screen editing events.
On April 13, 2024, Zhang Yong's team from Southwest University in China and his collaborators published a research paper titled "Boosting genome editing in plants with single transcript unit surrogate reporter systems" in Plant Communications. This research work is based on the previously constructed single transcript unit CRISPR-Cas (STU-CRISPR) genome editing system and constructed the STU-SR plant genome editing system based on the surrogate reporter system, which achieved efficient enrichment of Cas9 knockout editing, C to T and A to G base editing events, effectively expanded the plant genome editing toolbox, and provided strong technical support for precise plant genetic improvement.
The researchers built the STU-SR-SSA system based on the STU-CRISPR system, combined with a screening marker reporter gene containing endogenous gene editing sites. By using the same sgRNA to edit both the reporter gene and the endogenous gene simultaneously, a direct connection between reporter gene editing and target gene editing is established. When the reporter gene is cleaved by sgRNA, it can be repaired into a functional screening marker gene through SSA, thereby greatly increasing the probability of editing in the selected resistant plants. Experimental results in rice show that the STU-SR-SSA system achieved 100% editing efficiency at all tested endogenous gene loci. Compared with the traditional system, the editing efficiency was significantly improved. In addition, the biallelic editing efficiency has also been improved, with the biallelic editing efficiency of both OsPDS-sgRNA02 and OsDEP1-sgRNA01 reaching 100%.
In order to achieve effective enrichment of base editing events, researchers constructed the STU-SR-BE system. The reporter gene is obtained by removing the start codon of the marker gene and adding the endogenous gene site sequence. After the reporter gene is edited by C to T or A to G, a new start codon is formed and the normal function of the marker gene is restored. The results in rice show that compared with the traditional CRISPR-Cas9 system, the STU-SR-BE system significantly improves the editing efficiency on multiple endogenous gene sites, and combined with SpRY effectively expands the application scope of the STU-SR-BE system.
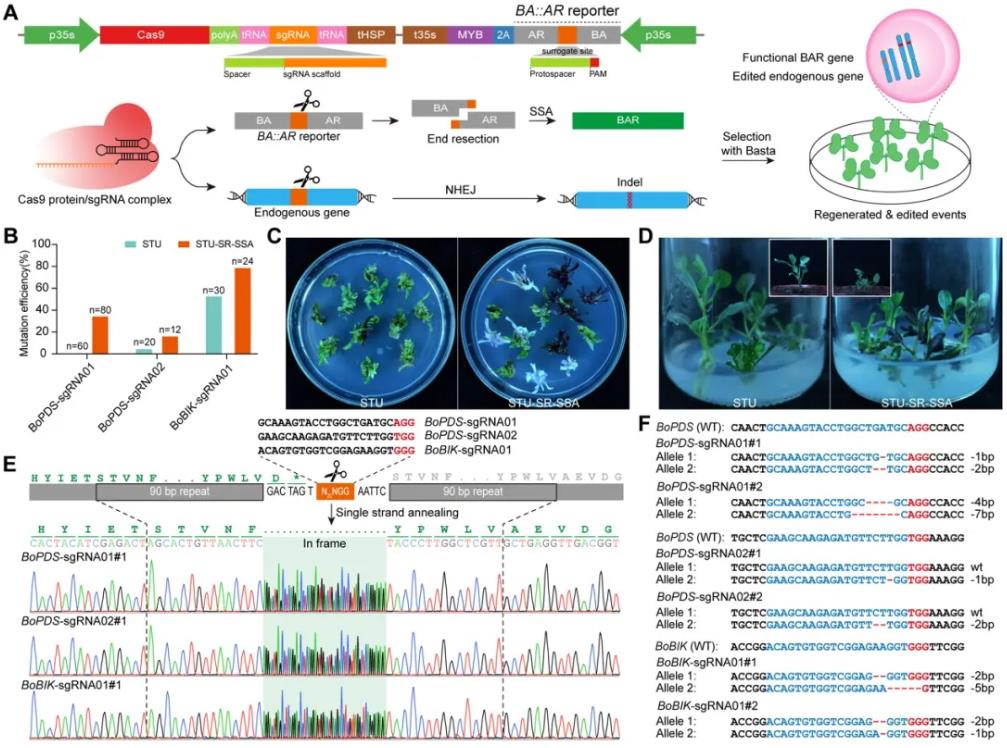
Finally, the researchers selected the dicotyledonous plant cabbage to test the applicability of the STU-SR system. Experimental results showed that compared with the traditional CRISPR-Cas9 system, the STU-SR-SSA system achieved higher editing efficiency on all tested endogenous gene loci. The editing efficiency of BoPDS-sgRNA01 site increased from 0% to 35%, the editing efficiency of BoPDS-sgRNA02 site increased from 5% to 16.7%, and the editing efficiency of BoBIK-sgRNA01 site significantly increased by 48.4%. This demonstrates that the STU-SR system can effectively enrich gene editing events and has broad application potential in different plant species.
| Cat# | Product Name | Price |
| CCVs-001 | pCas-Guide-Nickase | $704 |
| CCVs-002 | PX385 | $352 |
| CCVs-003 | pET-NLS-Cas9-6xHis | $352 |
| CCVs-004 | PX393 | $352 |
| CCVs-005 | PX398 | $352 |
| CCVs-006 | PX390 | $352 |
| CCVs-007 | PX392 | $352 |
| CCVs-008 | pFGC-pcoCas9 | $352 |
| CCVs-009 | pGuide-IT-ZsGreen1 | $2,816 |
| CCVs-010 | PX395 | $352 |
Reference