Insect Hi-C Assisted Genome Assembly
Hi-C (high-throughput chromosome conformation capture) technology is a technique used to study the three-dimensional conformation of the entire genome and analyze interactions between chromatin fragments, which can be used to reveal interactions within the genome. Lifeasible specializes in insect research and leverages its robust Hi-C technology to contribute to the advancement of insect genomics.

Advantages of Hi-C in Assisting Genome Assembly
- Hi-C does not require a population, it can achieve chromosome localization using a single individual. Hi-C determines chromosome localization based on differences in interaction frequencies caused by variations in spatial and linear distances on chromosomes, thus eliminating the need to construct a population.
- Higher marker density and more complete sequence localization. Compared to genetic maps, the interaction frequency between chromatin regions exhibits higher marker density. Using Hi-C technology, over 90% of genomic sequences can typically be localized to chromosomes.
- Correcting errors in assembled genomes. Hi-C data can be used to identify assembly errors, remove redundant sequences, and enhance genomic accuracy.
What Can We Offer?
Based on Hi-C technology, we provide one-stop services for insect genome research, from data processing to result evaluation, including the following service contents.
| Data statistics and filtering |
- Perform quality control on insect Hi-C sequencing data and remove low-quality and contaminated sequences.
- Ensure that the data used for assembly is clean and reliable.
|
| Hi-C library evaluation |
- Evaluate the quality of Hi-C library construction, check the insertion fragment length distribution, and the effective pairing ratio.
- Ensure the accuracy of subsequent chromosome positioning.
|
| Hi-C chromosome positioning and assembly |
- Grouping. Divide the assembled contigs/scaffolds into corresponding chromosome groups.
- Group sorting. Sort the sequences according to Hi-C contact frequency.
- Intra-group orientation. Use Hi-C signals to determine the orientation of sequences, improving chromosome assembly accuracy.
|
| Post Hi-C mapping statistics and evaluation |
- Reference genome evaluation for closely related species. Evaluate the reasonableness of assembly results by aligning with closely related insect genomes.
- Genetic marker evaluation. Verify chromosome assembly accuracy using genetic maps and SNP markers.
|
We have extensive experience in insect Hi-C-assisted genome assembly and offer customized solutions and technical support. Our insect genome assembly service effectively improves assembly efficiency and lays a solid foundation for subsequent functional annotation, evolutionary analysis, and other research.
Application Areas of Hi-C in Assisting Genome Assembly
- Chromosome-level genome assembly. Insect genomes are typically complex and contain many repetitive sequences. Hi-C can help achieve de novo assembly at the chromosome level.
- Genome error correction and optimization. Hi-C data can reveal assembly errors, such as misaligned or duplicated fragments, which helps improve the accuracy of the genome.
- Studying chromatin's three-dimensional structure. Revealing topological domains (TADs), A/B compartments, and other features of the genome helps to understand gene regulatory mechanisms.
- Species evolution and comparative genomics. By comparing chromosome structures, it is possible to study evolutionary relationships and gene family expansions among insect species.
- Assisting in functional gene localization. In studies of insect drug resistance, developmental regulation, behavior, and other areas, Hi-C assists in localizing functional genes, improving research efficiency.
Our Testing Process
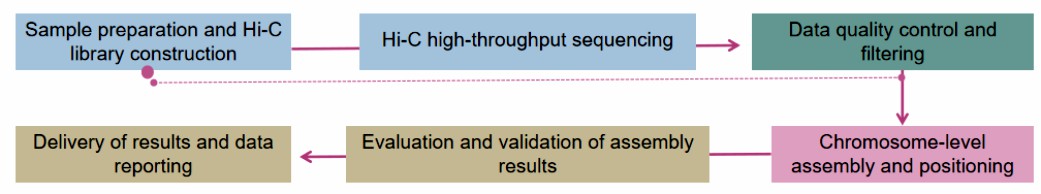 Fig.2 Our experimental process. (Lifeasible)
Fig.2 Our experimental process. (Lifeasible)
Why Choose Us?
- Stable Hi-C library construction system. We ensure high-quality Hi-C libraries.
- Independently developed process. We have established a Hi-C error correction process and iterative assembly process that can achieve a chromosome mounting rate of over 95%, detect assembly errors, remove redundant sequences, and even achieve monotype assembly, thereby improving genome quality.
- Extensive project experience. We have extensive experience in Hi-C-assisted genome assembly for various insect genomes.
Lifeasible specializes in insect research and has accumulated extensive experience in Hi-C-assisted genome assembly for insects. We continuously optimize our service system and update our technical platform to ensure the success of your scientific research. If you are interested, please feel free to contact us.
For research or industrial raw materials, not for personal medical use!

 Fig.2 Our experimental process. (Lifeasible)
Fig.2 Our experimental process. (Lifeasible)