PacBio Full-Length Transcriptome Sequencing of Insects
PacBio full-length transcriptome plays an important role in helping researchers understand the biological functions of insects. Based on the PacBio third-generation sequencing platform, transcriptome research can directly read the full-length cDNA without interruption, enabling the effective acquisition of the entire sequence of high-quality single RNA molecules. Lifeasible focuses on the field of insects, and we aim to provide advanced technical methods and comprehensive solutions for insect research. We offer insect PacBio full-length transcriptome research services, utilizing third-generation sequencing technology to perform high-precision, long-read sequencing of insect RNA, thereby obtaining complete transcript information to reveal insect gene expression, splicing variations, fusion genes, and more.

Advantages of Insect PacBio Full-Length Transcriptome Sequencing Technology
- Overcomes the limitations of second-generation sequencing, such as short read lengths and incomplete information.
- Sequences full-length variants from the 5' end to the 3' end without the need for fragmentation and assembly, directly yielding complete information on full-length transcripts.
- High data accuracy eliminates the need for error correction, enabling precise localization of fusion genes, accurate characterization of splicing sites, and the discovery of new genes and variants.
What Can We Offer?
Second-generation sequencing technology cannot provide a large number of long transcripts and loses important information, such as alternative splicing, which greatly limits the in-depth utilization of transcriptomic data. The PacBio full-length transcriptomic sequencing technology we provide for insect research is designed to help researchers obtain longer and even full-length transcriptomes.
| Sample and RNA processing |
- Diversified processing of insect samples. We provide sample collection and pre-processing services for different developmental stages, different tissues, or specific experimental treatment groups.
- High-quality total RNA extraction. We provide removal solutions for interfering substances such as polysaccharides, polyphenols, and chitin in insects.
- RNA integrity testing.
- mRNA enrichment or full-length cDNA synthesis.
|
| Insect PacBio full-length transcriptome sequencing |
- cDNA library construction.
- Iso-Seq sequencing performed on the PacBio sequencing platform.
|
| Data analysis services |
- Raw data quality control and filtering.
- Full-length transcript identification.
- Transcript clustering and assembly.
- Alternative splicing event detection.
- Homologous gene and novel gene prediction.
- Transcription factor prediction and functional classification.
- Gene structure annotation optimization.
- Fusion transcript detection.
- Isoform quantification analysis.
|
| Advanced analysis and integration |
- Comparison with insect reference genomes and functional annotation.
- lncRNA, circRNA, and miRNA target prediction.
- SSR (simple sequence repeat) and alternative splicing site development.
- Differential expression analysis (combined with RNA-Seq data).
- Insect-specific gene family analysis.
- Gene regulatory network construction.
|
Application Areas of Our Services
| Molecular genetics and genome annotation |
- Accurate identification of simple repetitive sequences (SSRs) and single-nucleotide polymorphism (SNPs) sites in insect transcripts provides a molecular marker basis for population genetics and evolutionary research.
- Full-length transcript alignment with the genome accurately maps gene boundaries, alternative splicing sites, and new transcripts, significantly improving the accuracy and completeness of insect genome annotation.
|
| Growth and development regulatory mechanisms |
- Comparing transcripts from different developmental stages or tissue locations to identify key genes and alternative splicing events regulating insect development.
- Capturing full-length isoforms of development-related genes and combining them with expression quantification analysis to elucidate functional differences of genes during development.
|
| Environmental adaptation and stress resistance mechanisms |
- Investigate the transcriptome response of insects under stress conditions, identify alternative splicing patterns, and expression regulatory networks of stress-related genes.
- Precisely identify new genes and isoforms related to resistance, such as splicing variants of key enzymes in pesticide metabolism pathways, to reveal the molecular basis of resistance formation.
|
| Behavioral ecology and biological interactions |
- Study transcript differences associated with insect foraging, migration, and other behaviors, and identify alternative transcripts and non-coding RNAs that regulate behavior.
- Analyze gene expression changes in insect interactions with microorganisms or host plants, and reveal interaction-specific transcripts.
|
Sample Requirements
| Sample type |
Insects |
| Recommended sample quantity |
No less than 8 specimens |
| Transportation method |
Dry ice transportation |
| Storage method |
Long-term storage at -80°C |
Our Testing Process
- mRNA enrichment. Enrich polyA-tailed mRNA.
- Reverse transcription. Transcribe into full-length cDNA.
- PCR amplification. Enhance the signal and add barcodes.
- Library construction. Repair the ends and connect adapters.
- Sequencing. Use the PacBio platform.
- Data analysis. Identify full-length transcripts.
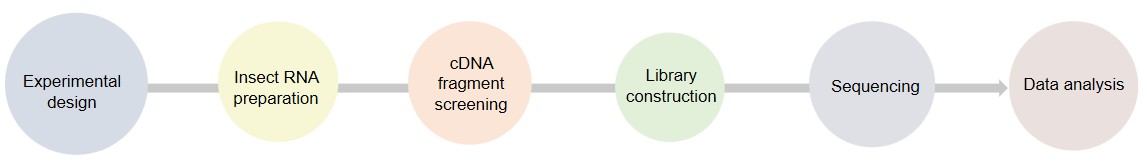 Fig.2 Our experimental process. (Lifeasible)
Fig.2 Our experimental process. (Lifeasible)
Why Choose Us?
- We ensure high-quality testing data.
- We have extensive project experience.
- We provide comprehensive analysis content.
- We offer customizable project designs.
Lifeasible focuses on the field of insect research. We have introduced the advanced PacBio third-generation sequencing platform and have a complete set of nanopore sequencers of various models. We provide accurate and high-quality full-length transcriptome services for insects. If you are interested, please feel free to contact us.
For research or industrial raw materials, not for personal medical use!

 Fig.2 Our experimental process. (Lifeasible)
Fig.2 Our experimental process. (Lifeasible)