Small RNAs (sRNAs) are a class of important non-coding RNAs that regulate gene expression through multiple pathways. Small RNA (sRNA) sequencing can detect the expression levels and expression differences of known and unknown miRNAs. When combined with transcriptomic sequencing data from the same sample for joint analysis, it can simultaneously analyze the expression of miRNAs and their target genes, providing a powerful tool for studying RNA function and regulatory mechanisms.
Lifeasible has extensive experience in research and services within the field of insects. Leveraging our advanced technical platforms and a team of expert professionals, we offer cutting-edge, specialized small RNA sequencing services tailored for insect samples. These services focus on high-throughput sequencing and analysis of small RNAs, and are widely applied in research areas such as gene expression regulation, developmental mechanisms, and immune responses.
In insect genomes, the transcription start sites of miRNAs are typically located in intergenic regions, introns, and the reverse complementary sequences of coding sequences. Their precursors have a typical hairpin structure, and Dicer/DCL enzymes generate the mature forms. For known miRNAs, we align the sequencing reads with mature miRNA sequences in the insect reference genome and specific databases to achieve precise identification. For unknown miRNAs, we use particular software for prediction, combined with insect-specific biological characteristics, to identify potential new miRNAs.
Using small RNA sequencing data, we can detect miRNA expression levels with high sensitivity. We quantify using TPM (Transcripts Per Million) and analyze density plots to visually display the overall expression patterns and dispersion of miRNAs in individual insect samples, as well as compare miRNA expression differences between insect samples under different experimental conditions or at different developmental stages.
Clustering analysis of differentially expressed miRNAs helps reveal dynamic changes in insect gene regulation under different experimental conditions. Hierarchical clustering methods group miRNAs with similar expression trends, enabling the inference of potential functions for unknown miRNAs or the identification of new roles for known miRNAs in specific physiological or metabolic processes in insects.
miRNAs bind to target gene transcripts through complementary pairing with their seed regions, thereby inhibiting or degrading corresponding mRNAs and regulating insect gene expression and phenotypic traits. We perform GO (Gene Ontology) functional annotation and KEGG (metabolic pathway) enrichment analysis on the target genes of differentially expressed miRNAs, and further elucidate the molecular mechanisms of miRNAs in insect development, metabolic regulation, immune responses, and environmental adaptation processes.
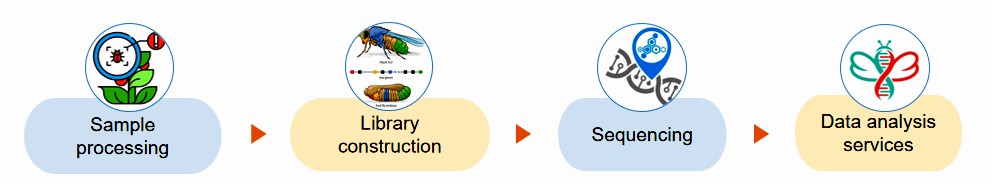 Fig.2 Our experimental process. (Lifeasible)
Fig.2 Our experimental process. (Lifeasible)
Lifeasible specializes in providing high-quality, accurate insect small RNA sequencing services to advance research in the field of entomology. If you are interested, please feel free to contact us.