Plant Secondary Metabolism Research Fills a Gap in the Iridoid Biosynthesis Pathway
On August 3, 2025, a collaborative team led by Maite Colinas and Sarah E. O’Connor from the Max Planck Institute for Chemical Ecology, along with C. Robin Buell from the University of Georgia, published a research paper titled “Discovery of iridoid cyclase completes the iridoid pathway in asterids” in Nature Plants. This landmark study in the field of plant secondary metabolism fills the final gap in the iridoid biosynthesis pathway.
Background and Significance
Iridoids are a class of monoterpenoid secondary metabolites widely found in asterids, possessing important ecological defense functions and medicinal properties. They are precursors to several important alkaloids, such as the anticancer drug vinblastine and the antimalarial drug quinine. Although the iridoid biosynthesis pathway has been extensively studied, the enzymatic mechanism of the key step, cyclization, has remained elusive.
Key Findings
This study successfully identified and functionally validated iridoid cyclase (ICYC) through single nucleus RNA sequencing (snRNA-seq), tissue-specific co-expression analysis, and heterologous expression systems. This enzyme, a member of the MES-type α/β hydrolase family, catalyzes the cyclization of the unstable intermediate 8-oxocitronellyl enol into two major stereoisomers:
- 7S-cis-trans
- 7R-cis-cis nepetalactol
Technical Innovations and Highlights
Multi-omics Integration Strategy
- Combining genomic, transcriptomic, and snRNA-seq, the researcherspinpointed candidate genes co-expressed with known iridoid pathway genes.
- Using cell-type-specific expression maps, the researchersrevealed the spatial organization of iridoid biosynthesis in IPAP cells (inner phloem-associated parenchyma cells).
Functional Verification System
- The entire iridoid pathway was reconstructed in Nicotiana benthamianato validate the function of ICYC.
- In vitro enzyme activity assays and virus-induced gene silencing (VIGS)confirmed the necessity of ICYC for the cyclization reaction.
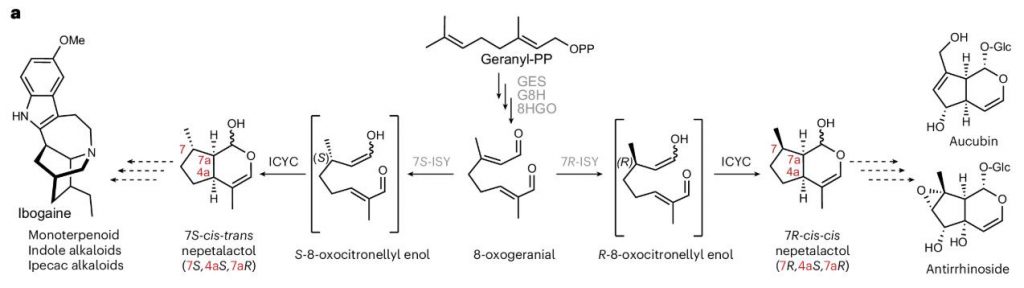
Figure 1. Iridoid biosynthesis includes previously characterized iridoid pathway genes. (Colinas, et al. 2025)
Evolutionary and Mechanistic Insights
- ICYC forms a conserved gene cluster with the G8H gene, suggesting that it may achieve metabolic channeling through co-localization.
- Unlike independently evolved cyclases (such as MLPL proteins) in Nepeta, ICYC represents a single-origin cyclization mechanism in Asteraceae.
Scientific Significance
- This study fills the final gap in the iridoid biosynthetic pathway, enabling a complete elucidation of the total synthesis pathway from GPP to various iridoids and their derivatives.
- This study reveals new functions for MES family enzymes and expands our understanding of the catalytic diversity of the α/β hydrolase superfamily.
- This study provides key enzymatic tools and a synthetic biology platform for metabolic engineering to produce high-value iridoid and alkaloids (e.g., anticancer and antimalarial drugs).
Summary
This study not only achieved breakthrough progress in plant natural product biosynthesis but also provided important tools and theoretical foundations for subsequent synthetic biology and drug development. Its multidisciplinary research strategy (genomics, enzymology, metabolomics, and synthetic biology) is worthy of reference and serves as a model for the integration of plant metabolic engineering and natural product chemistry.
Related Services
Reference
- Colinas, M., et al. (2025). Discovery of iridoid cyclase completes the iridoid pathway in asterids. Nature Plants, 1-13. DOI: 1038/s41477-025-02122-6.
For research or industrial raw materials, not for personal medical use!
