Chloroplasts are essential organelles that convert light energy into chemical energy in plants and are semi-autonomous organelles with a unique genetic system. The genes involved in photosynthesis in chloroplasts are relatively conserved in size, structure, and content. The chloroplast genome is mainly used to encode proteins and ribosomal proteins that are closely related to photosynthesis. Analysis of the regulation of chloroplast gene expression and genetic characteristics can help advance the study of the origin and evolution of plant species and the relatedness between different species.
Lifeasible has a long history of molecular plant research and can provide chloroplast genome sequencing services for many species. We can provide personalized and optimized solutions to difficult chloroplast sequencing problems. We are committed to providing our clients with accurate chloroplast genome sequencing services to help botanists conduct effective research on plant phylogeny, population evolution, and genetics.
We take the most suitable sequencing solution according to the different needs of our clients.
| Our successful project experience | ||
|---|---|---|
| liciaceae; Winteraceae | Drynariaceae | Dryopteridaceae |
| Plumbaginaceae | Zingiberaceae | Verbenaceae |
| Liliaceae | Asteraceae; Compositae | Magnoliaceae |
| Labiatae; Lamiaceae | Kareniaceae | Vitaceae |
| Leguminosae | Chenopodiaceae | Anacardiaceae |
| Poaceae; Gramineae | Prunoideae; Prunusoidea | Rosaceae |
| Cucurbitaceae | Polygonaceae | Solanaceae |
| Theaceae | Cruciferae; Brassicaceae | Schisandraceae |
| Sphagnaceae | Amaranthaceae | Chlorellaceae; Palmellaceae |
| Osmundaceae | - | - |
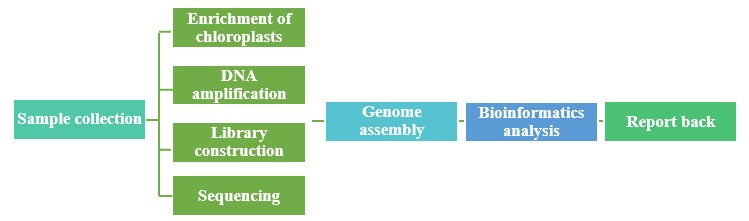
Lifeasible has recruited a large number of experienced sequencing technicians to form a team of professional plant sequencing technicians who can provide affordable, fast, and accurate chloroplast sequencing solutions to our clients. If you have any technical difficulties with your chloroplast genome research, please feel free to contact us.