Completing sequencing an increasing number of fungal genomes provides the conditions for revealing the function of fungal genes at the analytical level. However, the study of fungal genetics is seriously hampered by the relatively large fungal genomes, complex cell structure, lack of sexual stages in most fungi, and difficulties in gene manipulation. Lifeasible can establish fungal genetic transformation systems that provide for targeted genetic modification of fungal strains through insertion, deletion, and knock-down strategies.
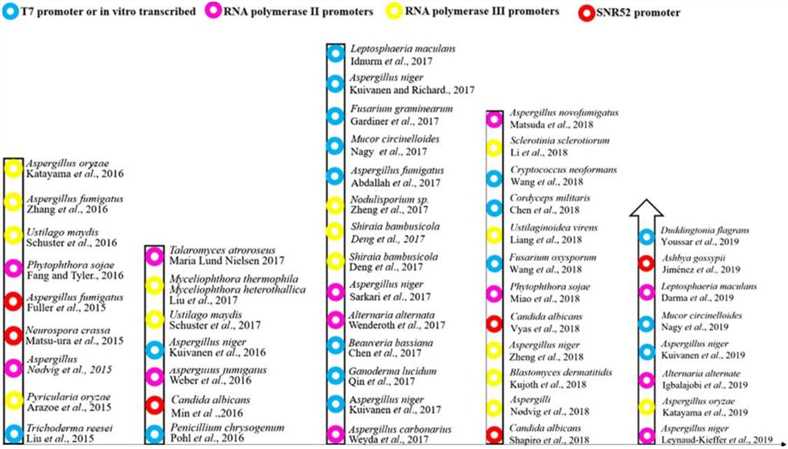 Figure 1. The history of the development and application of CRISPR/Cas9 technology in filamentous fungi. (Song R, et al., 2019)
Figure 1. The history of the development and application of CRISPR/Cas9 technology in filamentous fungi. (Song R, et al., 2019)
The CRISPR/Cas system consists of a manipulator encoding a Cas protein and a CRISPR repeat spacer sequence. A typical CRISPR repeat spacer sequence consists of a series of short, highly conserved forward repeats and spacer sequences of similar lengths in a certain order. Cas genes are a conserved family of genes that encode nucleases, DNA-decapping enzymes, polymerases, and other proteins related to the cleavage and modification of nucleic acids.
Genetic manipulation in fungi is often time-consuming and cumbersome, especially in diploid strains that lack a sexual cycle. Two core components of the CRISPR/Cas gene editing system include the Cas9 protein, which is used to generate DNA double-strand breaks, and the sgRNA, which is capable of directing the Cas9 nucleic acid endonuclease to any locus in a chromosome with a suitable prototypical spacer sequence adjacent to the motif.
The mechanism of action of CRISPR/Cas-mediated fungal gene editing consists of three main steps, which are the acquisition of new spacer sequences, tracrRNA synthesis and processing, and CRISPR/Cas9-mediated silent immune interference. When the constructed exogenous CRISPR/Cas system is introduced into the recipient cell, the sgRNA recognizes the target site, and Cas9 is responsible for the cleavage of the fungal DNA double strand; we can help our customers to introduce the target DNA fragment into the cell and realize the targeted editing of the target gene.
Lifeasible can use CRISPR/Cas9 gene editing systems to help customers advance their research related to fungal functional gene mining, targeted improvement of varieties, and targeted mutation of development-related genes. As your trusted partner, we can meet all your fungal phylogenetic analysis needs and provide you with efficient and high-quality services. If you want to know the details, please contact us.
Reference
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024
Application of Exosomes in Facial Beauty
April 12, 2024