Fungi are lower organisms with vast, and diverse species. Fungi exist widely in soil, air, and organisms inside and outside and have a very close relationship with human production and life. Some fungi, such as mushrooms, are directly used for food and medicine or have an essential role in the fermentation industry, food processing industry, and antibiotic production. Some fungi can cause diseases in plants. Therefore, the rational use of beneficial fungi and the control and prevention of harmful fungi are of great importance.
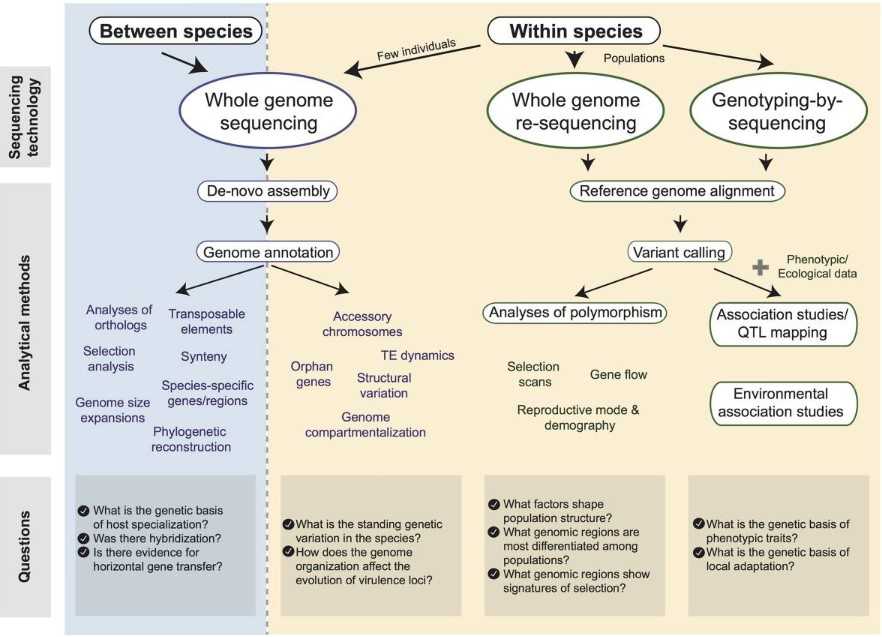 Figure 1. Diagram of tools available for the analysis of fungal pathogen genomes. (Plissonneau C, et al., 2017)
Figure 1. Diagram of tools available for the analysis of fungal pathogen genomes. (Plissonneau C, et al., 2017)
The fungal genome is relatively simple compared with other eukaryotes, easier to be sequenced and annotated, and easy to genetic manipulation and gene modification. Studying fungal genomics can help further understand the physiological properties, morphological differences, and evolutionary and metabolic diversity of fungi in different habitats. Lifeasible offers high-throughput gene sequencing-based genome analysis services for biotrophic, hemibiotrophic, and necrotrophic fungi.
We can provide both fungal genome de novo sequencing and fungal genome resequencing methods to obtain whole genome sequence information of fungi. By assembling and annotating fungal genome sequences and using bioinformatics analysis, we can study the molecular biological basis of fungal pathogenesis, host-host interaction mechanism, and metabolic mechanism.
The development of molecular biology techniques and their integration with systematics have revolutionized the study of fungal systematics. We are able to provide fungal phylogenetic analysis based on small ribosomal subunit, ribosomal large ribosomal subunit, internally transcribed spacer, and protein-coding gene sequences.
Compared with traditional population genetics, many molecular genetic markers can more efficiently infer the genetic structure and genes of plant pathogenic fungal populations. We can analyze the diversity and structure of fungal communities at the kingdom level, phylum level, class level, order level, family level, and genus level by sequencing large ribosomal subunits, small ribosomal subunits, internal transcribed spacer regions, or by macrogenome sequencing.
Comparative population genomics can provide a better understanding of pathogen origins, species formation, emergence, host tracks, and host hopping. It can also better capture the impact of hybridization, horizontal transfer, and chromosomal rearrangements on the emergence of new pathogens. We can compare fungal group DNA sequences and elucidate fungal genome diversity by performing family clustering analysis of fungal genes, species divergence time estimation analysis, and gene family contraction and expansion analysis.
Pan-genomics is the deep sequencing and de novo assembly of different individuals of a species to distinguish between the core genome, non-essential genomes, and individual-specific genes of the species. Fungal pangenomes can be used to obtain complete variation information of a large number of strains, which is helpful for fungal domestication and genotype-phenotype association studies. We can analyze the structural variation and functional gene localization of fungal pangenomes through fungal pangenome construction and sequence annotation.
Fungal genotyping is a technique that uses biological methods to determine an individual's genotype. For fungal genotyping, we can analyze fungal genotypes by ribosomal DNA, microsatellite polymorphism sequencing, etc., which provides a common basis for fungal gene function research, pathogenic gene identification, etc.
Lifeasible can provide fungal genome analysis services based on fungal gene sequencing, including fungal phylogenetic analysis, community structure analysis, comparative genome analysis, pangenome analysis, and genotyping. As your trusted partner, we can meet your fungal phylogenetic analysis needs and provide you with efficient, high-quality services. If you want to know the details, please contact us.
Reference
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024
Application of Exosomes in Facial Beauty
April 12, 2024