The Electrophoretic Mobility Shift Assay (EMSA) is a core technology used to detect the interactions between protein (complexes) and nucleic acids (1). This assay is widely applicable to various bio-sample analyses, and is capable of delivering both qualitative and quantitative characterizations of nucleic acid-protein interactions (2). Lifeasible provides services for sensitive, rapid EMSA assays that can examine interactions among a wide range of plant proteins and nucleic acids.
The principle of the EMSA assay is based on the electrophoretic mobility changes upon nucleic acid-protein interactions. Specifically, during gel electrophoresis, the motilities of protein-DNA or protein-RNA mixture vary depending on their size, electric charge and/or stoichiometry structure. Compared to the unbound DNA or RNA fragments, those that bind with a protein or protein complex would move more slowly, represented by a separated band indicating a larger size on the gel (Figure 1) (3). Further, the nucleic acids can be detected via radioisotope labeling, fluorescence labeling, chemiluminescence or immunohistochmical detection methods.
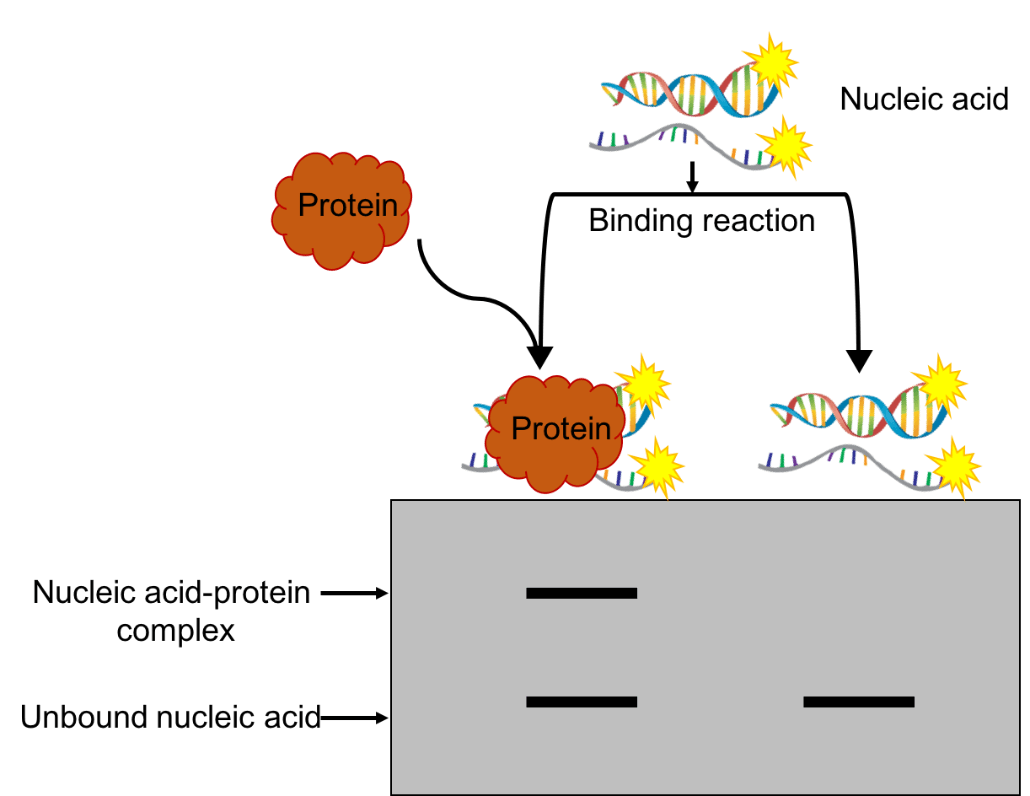 Figure 1. A schematic representative of the classic EMSA.
Figure 1. A schematic representative of the classic EMSA.
Compared to other protein-nucleic acid interaction assays, the EMSA holds the following advantages:
Aside from the classical EMSA, Lifeasible also offer a number of alternatives, such as nitrocellulose filter-binding and footprinting (4, 5), that are optimized to allow quantitative detections of binding kinetics as well as equilibrium measurements. Here at Lifeasible, our team of skilled scientists and experts has years of experience in both classical EMSA and other alternative EMSA assays. Our quality-ensured and technically flexible services make sure that your needs for each project can be met.
References
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024
Application of Exosomes in Facial Beauty
April 12, 2024