
Plant diseases affect normal growth and development and can even lead to plant death. Most plant diseases are caused by pathogenic microorganisms or viruses. Bioinformatics analysis of plant metagenome can reveal information about the microbiome in plants, which is critical for pathogen identification and species diversity analysis. With the development of sequencing technology, metagenomic has become a current research hotspot and is one of the important tools for studying plant diseases.
As a leading plant sequencing company, Lifeasible has extensive sequencing equipment and a professional bioinformatics analysis team to provide one-stop bioinformatics analysis solutions for plant metagenome for universities and research institutions. We strive to meet all our customers' needs by constantly optimizing our test protocols and improving our service system.
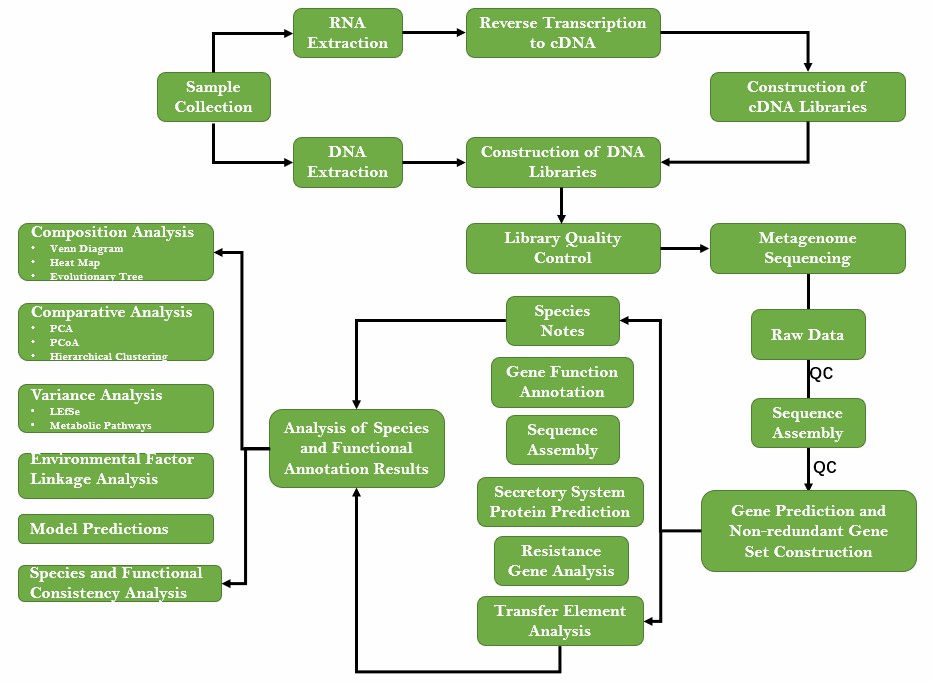
We extract nucleic acids from all microorganisms in a given environment, construct a sequencing library, sequence the data, and then quality control, assemble, compare and annotate the data to obtain all microorganisms in a given environment or design primers based on the rDNA database, and finally obtain information on their biodiversity and gene function with the help of bioinformatics. The specific services are as follows.
We use long-read technology to determine the full range of microbial genetic information in a given environment.
The sequenced base sequences were quality-controlled using software such as FastQC and KneadData, including the removal of sequencing junctions, removal of hosts, and removal of low-quality bases.
The high-quality sequences obtained are analyzed with or without reference using software such as Kraken2 or MEGAHIT. In the case of reference genomes, all sequenced fragments are first positioned on the reference genome by sequence alignment, and those that match well are selected for subsequent analysis; in the case of non-reference analysis, there is no reference genome, and the assembly results are generally contigs.
Prokka is then used for gene annotation and species classification. For the annotated genome, non-redundant gene sets can be constructed to describe the overall information of genes in the environment.
Lifeasible is dedicated to providing plant biologists with plant metagenome bioinformatics analysis services that meet their needs. If you have questions, please contact us directly.
Reference