Single Nucleotide Polymorphism (SNP) is a DNA sequence polymorphism caused by the variation of a single nucleotide at the genomic level. SNPs include single base conversions, reversals, insertions, and deletions. SNPs are the third generation of DNA-level genetic polymorphism markers after RFLP, VNTR, and STR.
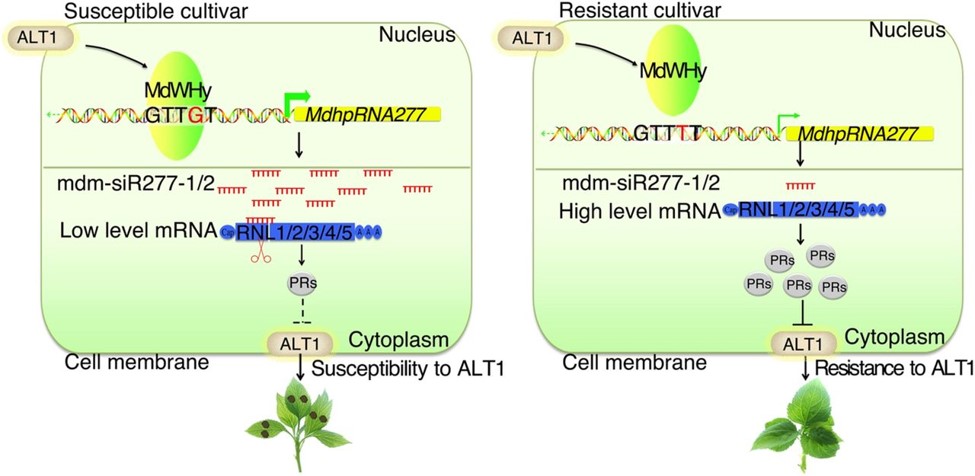 Figure 1. Model for the Role of SNPs in pMdhpRNA277 in Regulating A. alternata Leaf Spot Resistance in Apple. (Qiu lZ, et al. 2018)
Figure 1. Model for the Role of SNPs in pMdhpRNA277 in Regulating A. alternata Leaf Spot Resistance in Apple. (Qiu lZ, et al. 2018)
Lifeasible has advanced instrumentation and years of experience in plant molecular biology and can provide you with rapid and comprehensive SNP detection and mutation analysis services using Direct sequencing method, KASP typing method, LDR typing method, Multiplex SNaPshot SNP typing method, Taqman probe method, etc.
| Typing Method | Technical Characteristics |
|---|---|
| Direct sequencing method | The highest accuracy, "gold standard", large workload, suitable for a small number of samples, many points, and centralized typing detection. |
| KASP typing method | It is suitable for typing detection with a large sample size and few loci. |
| ligase detection reaction (LDR) typing method | It is suitable for any polymorphic site, and there is no need to introduce restriction sites artificially. Accurate typing (> 95%), high detection rate, short cycle, and high performance-to-price ratio. |
| Multiple SNaPshot SNP typing method | The typing is accurate (> 98%), and multiple sites can be detected at the same time, which is not limited by the polymorphic characteristics of SNP loci and the number of samples. |
| Taqman probe method | The use of an MGB probe can greatly improve the signal-to-noise ratio and is suitable for detection with a small number of sites and high sample flux. The operation is simple, only a one-step PCR reaction is needed, no purification and pretreatment are needed, and the detection is directly on the computer with high accuracy. |

Lifeasible offers complete, professional SNP technology service, as well as customized experimental protocols based on your project requirements and plant sample characteristics. For more information, please contact Lifeasible.
Reference:
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024
Application of Exosomes in Facial Beauty
April 12, 2024