Detection of Epigenetic Modifications in Plants
Epigenetic modifications refer to heritable genomic alterations resulted from DNA modifications or chromatin structural changes, rather than DNA sequence changes. With the potential of altering DNA accessibility and chromatin organizations, epigenetic modifications play important roles in the regulation of gene expression.
In general, there are two types of epigenetic modifications, which often work collectively to regulate gene expressions:
- DNA methylation. This modification is catalyzed by DNA methyltransferase, which covalently attaches a methyl group to the C5 position of cytosine residues in CpG dinucleotide sequences.
- Histone modifications. Modification of histones can directly lead to alterations of chromatin structures. Two of the mostly recognized histone modifications are:
- Acetylation. Catalyzed by histone acetyltransferases (HATs), an acyl group is added to the amino acid of histone proteins, causing loosening of chromatin and promotion of gene activation. The acetylation modification can be reversed by histone deacyltransferases (HDACs).
- Methylation. Catalyzed by histone methyltransferases (HMTs), one or multiple methyl groups are transferred from S-adenosyl-L-methionine to lysine or arginine residues. Histone methylation can be associated with either activation or inhibition of gene expressions.
The phenomenon of epigenetic modification universally exists in eukaryotes, especially in plants. So far, there are over one hundred epigenetic regulators identified in the plant system. Noticeably, accumulating evidences have shown that epigenetic regulations contribute largely to the phenotypic diversity of plants, and more importantly, in the regulation of plant growth, development, and response upon environmental stimulations. Therefore, detection and characterization of epigenetic modifications are critical aspects for the study of plant biology.
Lifeasible is a leading plant biotechnology company with a long history in plant epigenetic research. We proudly provide our customers with multiple cutting edge technologies for the detection of epigenetic modifications, including:
- Detection methods for DNA methylation:
- Chromatin immunoprecipitation (ChIP)-based methods. Methylated regions of the genome are purified via methylation-specific antibodies. The isolated DNA fragments are then identified by microarray (ChIP-chip) or by next-generation sequencing (NGS) (Figure 1).
- Bisulfite modification of DNA. Bisulfite treatment specifically converts nonmethylated cytosines to uracils, which are then converted to thymines by PCR, while methylated cytosines are untouched. Bisulfite-treated DNA can then be analyzed by bisulfite sequencing, methylation specific PCR (MS-PCR) or methylation-based microarrays.
- Endonucleases digestion. By digesting of genomic DNA with endonucleases that differ in their methylation sensitivities, a rough estimate of the totality of methylation can be obtained.
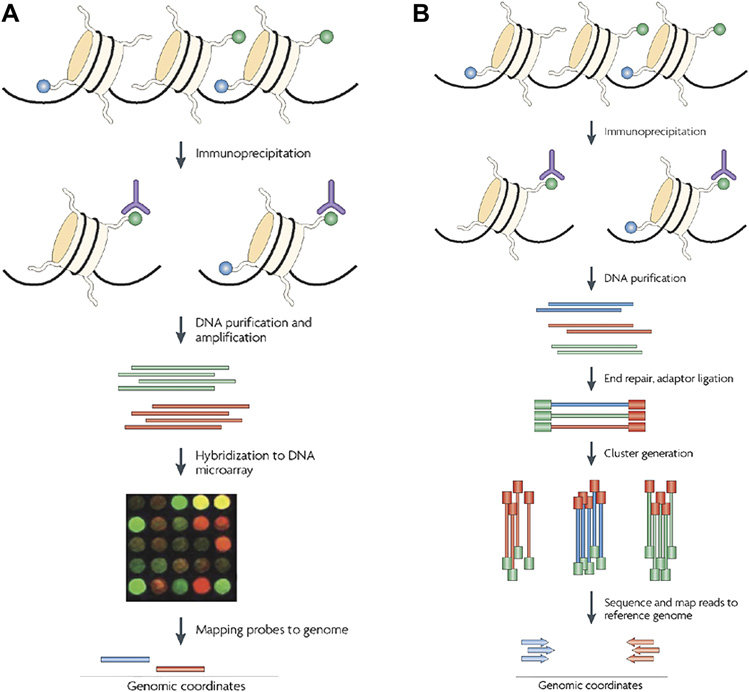 Figure 1. Illustrations of ChIP-based methods, ChIP-chip (A) and ChIP-seq (B)(Ku, Naidoo et al. 2011).
Figure 1. Illustrations of ChIP-based methods, ChIP-chip (A) and ChIP-seq (B)(Ku, Naidoo et al. 2011).
- Detection methods for chromatin modifications:
- Mass spectrometry. The mass spectrometry technique allows large-scale quantitative analysis of histone post-translational modifications and combinations.
- Chromatin immunoprecipitation (ChIP)-based methods. ChIP assays allow the analysis of chromatin structure surrounding specific DNA sequences (Figure 1).
- DNaseI hypersensitivity assays. As DNaseI hypersensitivity sites are usually located nearby the promoter regions, DNaseI hypersensitivity assays allow mapping of transcriptionally active versus inactive chromatin.
Lifeasible is devoted to providing our customers with services of the highest quality. Our experienced scientists and experts are always happy to help you with project designing, experimental optimization, data interpretation, technical consulting and so on. Welcome to contact us for questions, inquiries or collaborations.
Reference
- Ku, C. S., N. Naidoo, M. Wu and R. Soong (2011). "Studying the epigenome using next generation sequencing." J Med Genet 48(11): 721-730.
For research or industrial raw materials, not for personal medical use!
 Figure 1. Illustrations of ChIP-based methods, ChIP-chip (A) and ChIP-seq (B)(Ku, Naidoo et al. 2011).
Figure 1. Illustrations of ChIP-based methods, ChIP-chip (A) and ChIP-seq (B)(Ku, Naidoo et al. 2011).