Crop improvement has been the basis for food security for a growing population. Plant breeding works by exploiting genetic variation among individuals within a plant species and combining desired characteristics into new and improved varieties. Genetic variation is the variation in gene sequences between individuals in a plant population. Allelic variation is the basis of genetic variation and is expressed through different phenotypes. Processes such as mutation, random mating and fertilization, and gene duplication may introduce new genes and alleles that increase genetic variation. Genetic variation provides the basis for plant genetic diversity and reflects the degree/amount of variation present within a population. Without genetic variation, populations would not be able to adapt to different climatic conditions and could easily become extinct. In short, genetic variation is at the root of natural selection and is a crucial driver of the evolution of organisms. At the same time, the estimation of the extent and pattern of genetic variation is a prerequisite for the production of superior varieties. Lifeasible provides comprehensive plant genetic variation testing services to help you develop high-quality crop varieties for clients worldwide.
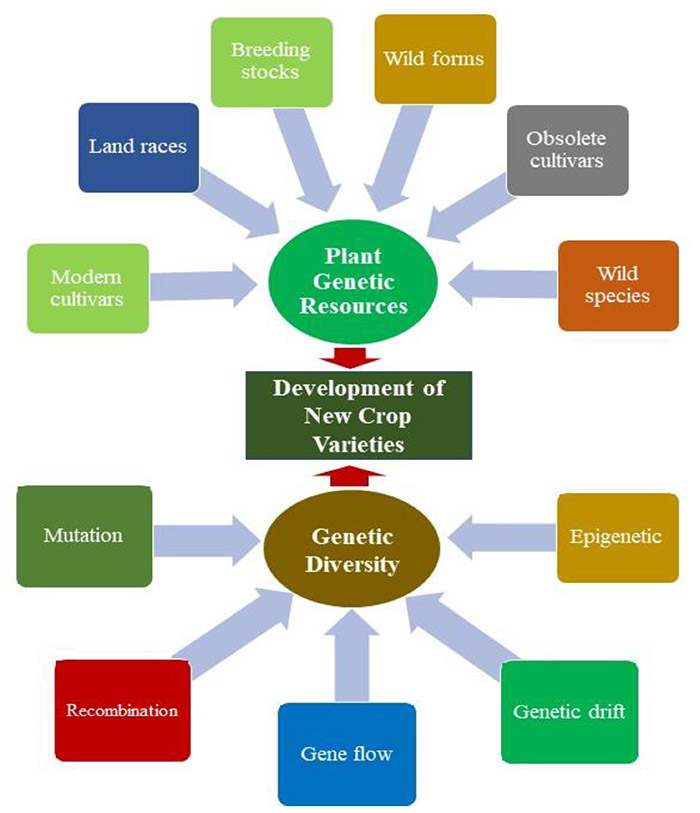 Fig. 1. Different sources of genetic diversity and their potential utilization in developing new crop varieties. (Salgotra R K et al., 2023)
Fig. 1. Different sources of genetic diversity and their potential utilization in developing new crop varieties. (Salgotra R K et al., 2023)
Genetic variation in plants is extensive and is a key factor in the adaptation of plants to different climates. There are many other methods used to amplify plant genetic variation, such as chemical or physical treatments, translocation breeding, synthetic hexaploidy, CRISPR/Cas9, etc. As a specialist in plant genetic engineering, Lifeasible provides professional solutions for detecting plant genetic variation, accurately detecting and analyzing both natural and epigenetic variation in plants. Our strict quality policy ensures the reliability of our results.
We offer various methods to detect the degree of similarity and difference between plant germplasm.
With high-throughput phenomics methods, we can provide detailed documentation of plant morphological traits, including characteristics from nutrient growth and reproductive growth to maturity stage. We ensure comprehensive and accurate data collection. In addition, we help you analyze complex traits such as yield and disease resistance by analyzing high-quality phenotypic data. This approach is easy, simple, inexpensive, and directly measurable.
By selecting markers related to morphological changes in chromosome size, shape, number, length, arm ratio, volume, cell division behavior, and DNA content. We offer Fluorescence In Situ Hybridization (FISH) and Genomic In Situ Hybridization (GISH) techniques to characterize foreign gene infiltration in plant species and to dissect the genetic composition of natural and artificial hybrids.
We offer isozyme marker technology to analyze and detect variation between and within plant species. This method is simple, fast, and inexpensive, making it ideal for projects that require low-level detection of genetic variation.
We offer a variety of molecular marker methods based on DNA sequence polymorphisms to detect plant genetic variation. Each marker system has its own advantages and disadvantages, and we will select the best solution for you. This approach offers the following advantages:
| Molecular Marker Method | Molecular Marker System |
| Markers based on nucleic acid hybridization | Restriction fragment length polymorphism (RFLP) |
| PCR-based markers | Random amplified polymorphic DNA (RAPD) Amplified fragment length polymorphism (AFLP) Microsatellite or simple sequence repeats (SSR) Randomly amplified microsatellite polymorphism (RAMP) Sequence-related amplified polymorphism (SRAP) Inter simple sequence repeats (ISSR) Target region amplified polymorphism (TRAP) |
| PCR-based markers | Cut amplified polymorphic sequences (CAPS) |
| Retrotransposon based markers | Inter retrotransposon amplification polymorphism (IRAP) Retrotransposon microsatellite amplification polymorphism (REMAP) Retrotransposon-based insertion polymorphism (RBIP) Inter primer binding site (iPBS) |
| Sequence-based markers | Single nucleotide polymorphisms (SNPs) |
| Array-based platforms | Diversity Array Technology (DArT) Restriction Site Associated DNA (RAD) Single Feature Polymorphism (SFP), etc. |
| Functional molecular markers (FMM) | Refer to DNA markers generated by sequence polymorphisms between functional genes associated with variation in a desired phenotypic trait. |
We offer a variety of sequencing technologies to comprehensively analyze plant genetic diversity and quickly identify useful genes and/or alleles.
Lifeasible provides comprehensive content of plant genetic variation detection and can design customized solutions according to your specific requirements. We hope that our products and technology can satisfy our customers. We look forward to your inquiry. For more information, please contact us.
Reference