According to the different functions and spatial distribution, the interior of cells can be further divided into other cellular regions or organelles, that is, subcellular. Each subcellular has a unique set of proteins, and the synthesized proteins can only function properly if they are guided by the correct protein sorting signals and transported to the appropriate subcellular, which is called plant protein subcellular localization. If the subcellular localization of plant proteins deviates, it can significantly impact plant cell function.
Lifeasible has established a leading plant proteomics platform, and we have provided you with plant protein subcellular localization services for a long time. We are confident in providing you with the most accurate and cost-effective solutions to help you explore the biological functions of plant proteins.
We design the most scientifically feasible project plan for you according to your research purpose and the characteristics of different plant subcellular. We can provide you with the following detection methods:
We have successfully provided our clients with various protein subcellular localization analyses in plants such as crucifers, monocotyledons, and valuable trees. These plants include but are not limited to, those shown in the table below.
| Our successful project experience | ||
|---|---|---|
| Oryza sativa L. | Nicotiana tabacum L. | Triticum aestivum L. |
| Hordeum vulgare L. | Zea mays L. | Arabidopsis thaliana |
| Pinus massoniana Lamb. | Areca catechu L. | Taraxacum kok-saghyz Rodin |
| Panicum virgatum L. | Sinomanglietia glauca | Dimocarpus longan |
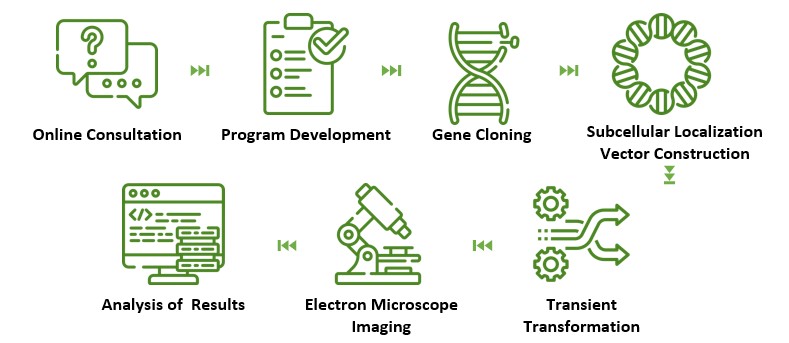
Lifeasible has built a comprehensive service platform for plant protein functional research and is committed to providing better scientific services to our clients. We look forward to working with you and providing quality plant protein subcellular localization services. If you have any interest, questions, or collaboration, please contact us directly.