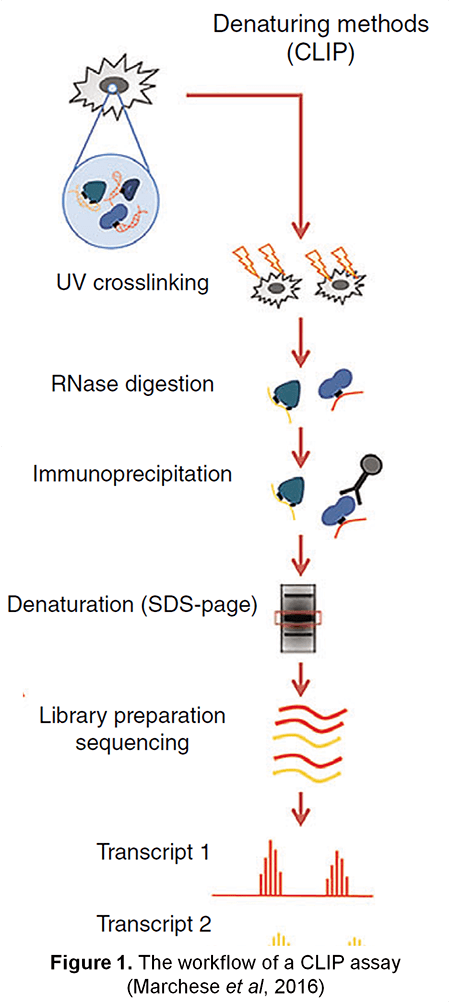
RNA binding proteins (RBPs) function in every aspect of RNA biology, including RNA transcription, pre-mRNA splicing and polyadenylation, RNA modification, transport, localization, translation, and turnover. Thus, development of technologies for studying RNA-protein interactions have been put under spotlight. Lifeasible, as a leader in plant biotechnology, has developed an advanced protein-centric method - cross-linking and immunoprecipitation (CLIP) - to map RNA binding protein binding sites or RNA modification sites in vivo under denaturing conditions. CLIP provides information on RNA-protein interactions on a genome-wide scale, promoting our understanding of post-transcriptional regulatory networks.
CLIP is performed by immunoprecipitation with a specific antibody against the protein-of-interest after UV cross-linking of the RNA-protein complex. UV irradiation induces covalent cross-linking only in sites where proteins directly interact with RNA. The treatment of RNases after UV exposure can obtain short RNA fragments and disrupt the RNA-mediated protein-protein interactions. The two steps above help reduce RNA-protein complexes by indirect bindings. After immunoprecipitation, the RNA-protein complexes are then separated from free RNA using sodium dodecyl sulphate -polyacrylamide gel electrophoresis (SDS-PAGE) gel and membrane transfer. Proteins of RNA-protein complexes are digested with proteinase K, leaving only a peptide at the cross-link site. This step allows for the identification of the cross-linked nucleotide. Finally, RNA fragments are reverse transcribed and analyzed by qRT-PCR or RNA-seq (Figure 1).
With excellent experts and rich experience in plant biotechnologies, we can design and develop customized CLIP protocols to meet customers’ needs. We pride ourselves on the short turnaround time and high-quality data. Moreover, we have also developed many advanced CLIP approaches, such as photoactivable ribonucleoside enhanced CLIP (PAR-CLIP), individual-nucleotide resolution CLIP (iCLIP), and cross-linking and analysis of cDNA (CRAC). These advanced CLIP methods are derived from modified cross-linking or library-preparation protocols of CLIP, allowing the identification of cross-linking sites at a single-nucleotide resolution (Figure 2). Please feel free to contact us for any further information on our services or assistance you might need.
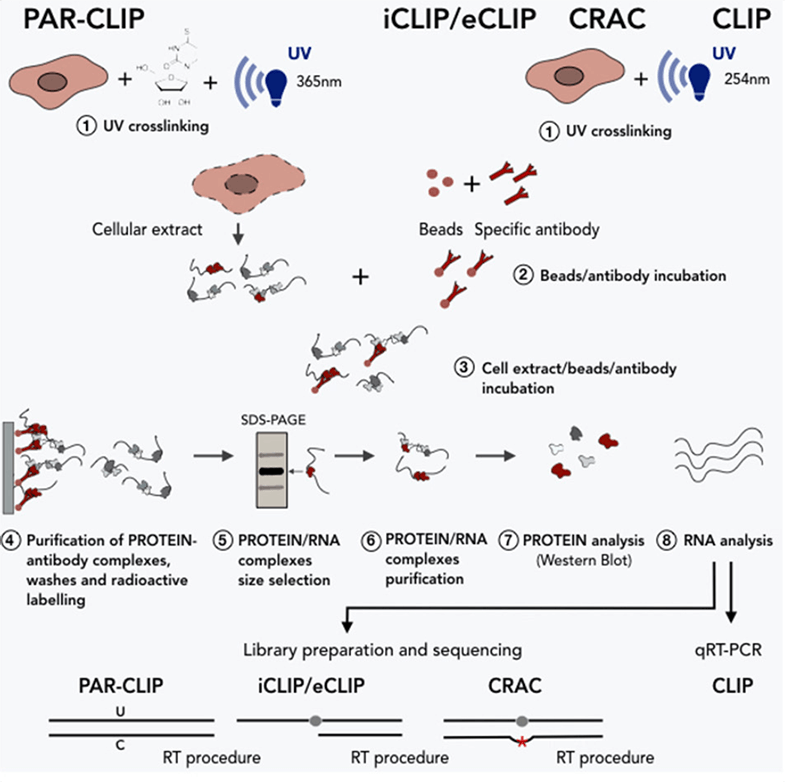 Figure 2. CLIP and advanced CLIPs for the study of RNA-protein interactions (Cipriano and Ballarino, 2018)
Figure 2. CLIP and advanced CLIPs for the study of RNA-protein interactions (Cipriano and Ballarino, 2018)
References