Lifeasible has been deeply involved in the field of gene editing for several years, and has accumulated a wealth of experience for relevant technologies. We are committed to optimizing the CRISPRa system to help customers achieve stronger activation effects.
Before the birth of CRISPR technology, there was no suitable technology to activate endogenous gene expression for biological community. No matter which method is used, it is necessary to obtain the cDNA sequence of the gene in advance, and the construction cost is relatively high, especially for large-scale cDNA library, which is very difficult to produce. The emergence of CRISPR technology allows us to activate gene expression in situ.
Through the modification of the CRISPR/dCas9 system, especially the modification of the guide RNA, the dCAS9 protein and the guide RNA can recognize the protein at the same time, and adsorb more transcription activators, thereby achieving a stronger transcription activation effect. The first generation of gene activation technology is about 7 times, while the second generation of gene activation technology can be as high as 40 times.
The third-generation gene activation technology mainly relies on the combination of multi-gene targeting technology and gene activation technology. This gene activation technology is very effective in dicotyledonous plants, By activating the flowering genes of Arabidopsis thaliana, the flowering time of Arabidopsis can be shortened from 40 days to 13 days, realizing strong activation of biallelic genes.

We are committed to optimizing the CRISPR system to carry more activation elements. After synergistic amplification, the CRISPRa system has a stronger activation effect and can really help our customers achieve gene transcription activation
We provide one-stop service, and customers only need to provide the gene information that needs to be activated. Our services cover gRNA design, vector construction, virus packaging and transfection, gRNA sequence screening, construction of CRISPRa stably transfected cell lines, target gene expression analysis, etc.
Among the many designs, the three technologies VPR, SAM and Suntag are the most popular ones.
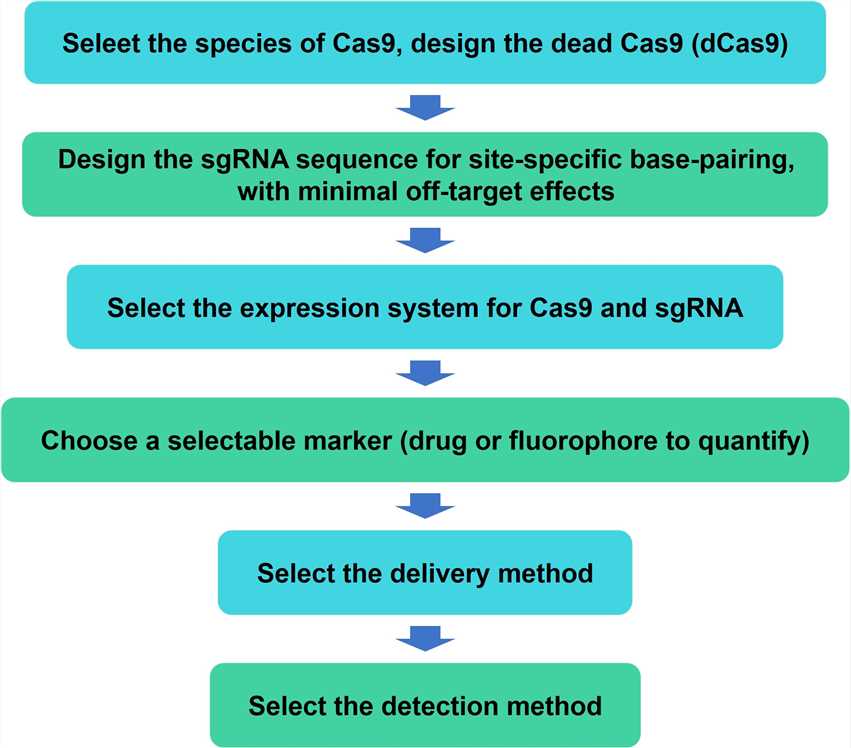
We hope to share our technology and results with customers to help customers advance their projects better and faster. For inquiries, please contact us.
01
Stronger activation effect
02
One-stop service to accelerate your research
03
Professional technical services
04
Experienced expert team
References
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024
Application of Exosomes in Facial Beauty
April 12, 2024