Lifeasible is a plant biotechnology company aiming for developing new varieties of field crops with high yield, great taste, long shelf-life, strong disease resistance and drought-tolerance. We provide specialized and customized one-stop services in association mapping for clients all over the world.
Most desired agricultural traits are polygenic and quantitative in nature and are controlled by collective effects of numerous genes on the same/different chromosome, known as quantitative trait loci (QTLs). Mapping of these QTLs in the genome of crop species using molecular markers is necessary for crop improvements. Linkage disequilibrium (LD)-based association mapping is an advanced QTL mapping method, which provides high mapping resolution with a large number of recombination events.
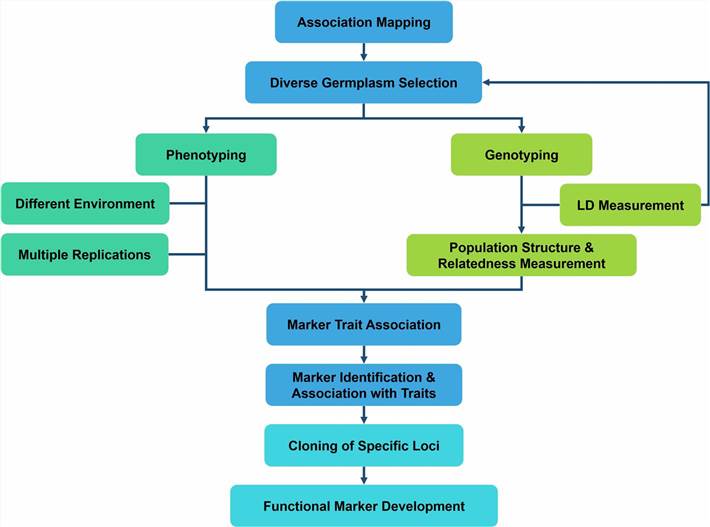 Figure 1. The methodology of Linkage disequilibrium (LD)-based association mapping (Nadeem et al., 2018)
Figure 1. The methodology of Linkage disequilibrium (LD)-based association mapping (Nadeem et al., 2018)
Being a team with strong knowledge of genetics, statistics, mathematics, and bioinformatics, Lifeasible is devoted to offering optimized and efficient QTL mapping and plant breeding services with reduced costs and maximal outcomes. The association mapping services we provide include:
We also have developed traditional biparental mapping (Linkage mapping) methods, which are more feasible for the genome-wide scanning for QTLs. We provide multiple optional mapping populations (such as double haploid lines (DHLs), backcross (BC) population, F2 population, F2:3/F2:4 lines, and recombinant inbred lines (RILs)) derived from two diverse parents for linkage mapping.
01
Optimized and efficient association mapping solution
02
One-stop service realizes seamless connection of research
03
Experienced team can meet individual needs
04
Shorten the breeding process for our customers
Welcome to contact us for technical consulting, experimental design and other informations; and our tech representatives are happy to walk you through our featured technologies, service plans, collaboration options and more.
In addition to providing professional association mapping services, Lifeasible also provides a variety of other breeding services to meet the needs of different customers. At the same time, we also provide plant genetic engineering, genetic transformation and analysis services.
Reference