As a prevalent and extremely critical post-translational modification of proteins in eukaryotic cells, ubiquitination can rapidly and specifically degrade abnormal proteins and regulatory proteins that transiently control a range of essential cellular life activities. Therefore, ubiquitination is important in plant growth and development, stress response, etc.
The process of protein ubiquitination modification involves a variety of enzyme systems, ubiquitin molecules, and substrate proteins. Ubiquitin activating enzyme (E1), ubiquitin-coupled enzyme (E2), and ubiquitin ligase (E3) are the key enzymes for protein ubiquitination modification. Detection of the biochemical properties and specificity of these ubiquitination-modifying enzymes and their substrate proteins is an essential element in analyzing their biological functions.
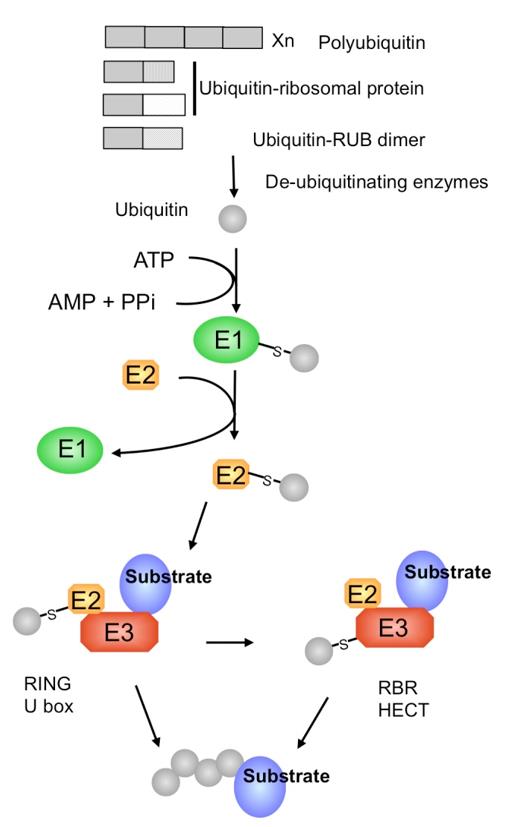 Figure 1. Ubiquitin genes and ubiquitination pathway. (Callis, J. 2014)
Figure 1. Ubiquitin genes and ubiquitination pathway. (Callis, J. 2014)
Lifeasible has been researching in the field of plant protein modification assay for many years. With its advanced technology platform, Lifeasible provides botanists with one-stop in vitro ubiquitination assay services for plant proteins from experimental design to data analysis, fully meeting the various research needs of botanists.
Lifeasible has developed a simple, rapid, and accurate in vitro ubiquitination assay to address the problems of high workload, complex reactions, and false negative rates of purified proteins expressed in vitro.
We cloned the target gene into the pET28a vector and transformed the plasmid into E. coli BL21 (DE3) or other suitable strains to express it in large quantities. After 24 h, the target protein with 6× His tag or MBP tag was extracted and separated, and the target protein was purified and desalted by ultrafiltration. Incubate with specific ubiquitin-modifying enzymes under the appropriate reaction buffer, time, temperature, and protein concentration. The incubated products were subjected to IP and WB analysis.
We transfect E. coli with the protein gene predicted to be vegetated and purify it after massive expression. Alternatively, the endogenous protein was extracted directly. They are incubated in vitro with K63- or K48-linked ubiquitin chain peptides and then detected by SDS electrophoresis with ubiquitinated antibodies. This method can clarify which lysine residues of the protein to be studied are susceptible to ubiquitination.
 Figure 1. Ubiquitin genes and ubiquitination pathway. (Callis, J. 2014)
Figure 1. Ubiquitin genes and ubiquitination pathway. (Callis, J. 2014)
The recognition of specific substrates is mainly determined by E3, and E2 regulates specific forms of ubiquitination modifications, such as monoubiquitination, multi-site monoubiquitination, and the generation of polyubiquitin chains of different configurations. Therefore, the combination of different E2-E3-substrates determines the specific form of ubiquitination modification of a specific protein, thereby affecting the fate of the substrate and regulating the physiological and biochemical pathways it participates in. We can provide you with the following services:
Lifeasible has established an effective ubiquitination detection system, which can provide high-quality in vitro ubiquitination assay services of plant proteins for researchers worldwide. For more information or any inquiry requirements, please contact Lifeasible.
Reference