In order to overcome the limitations of Drop-seq in the detection of single-cell RNA transcriptome, Lifeasible uses a more comprehensive and lower-cost method that can simultaneously perform RNA amplification sequencing and transcriptome analysis in a single cell- droplet-assisted RNA targeting by single-cell sequencing (DART-seq).
The droplet microfluidic technology has changed the development and application of single-cell sequencing technology, and provides a low-cost and high-throughput method for single-cell genomics research. In 2015, researchers collaborated to develop Drop-seq. This technology introduces microfluidic technology into the single-cell RNA-Seq method, using nano-grade droplets and attaching a unique barcode to the RNA of each cell. Drop-seq can effectively characterize the gene characteristics of thousands of single cells at the same time, making it a reality to analyze the genes of many single cells quickly and at a low cost.
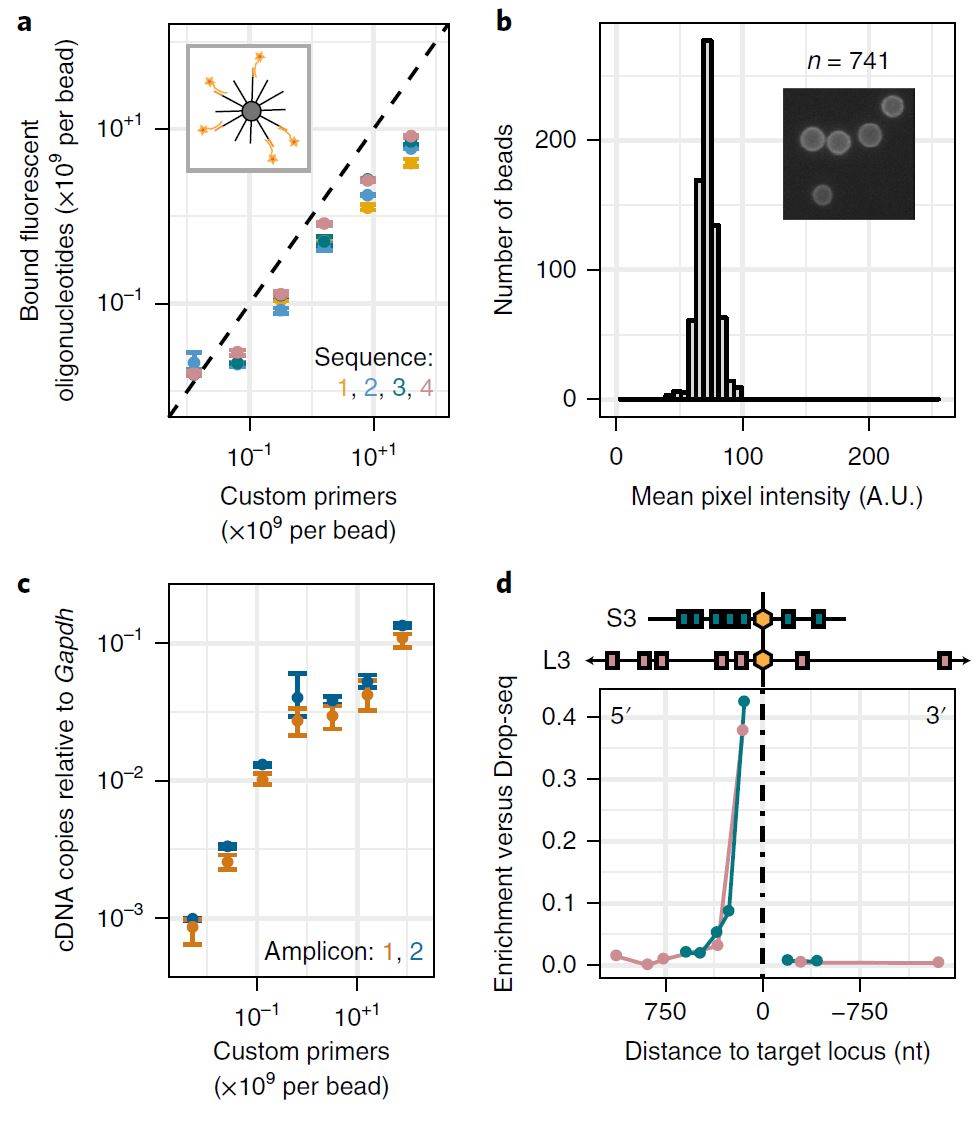 Figure 1. Characterization of DART-seq primer bead synthesis and comparison of DART-seq and Drop-seq. (a) Number of fluorescence probes bound per bead as a function of the number of primers per bead included in the ligation reaction (four distinct primer sequences). Schematic of fluorescence hybridization assay (inset), (b) Bead-to-bead variability in fluorescence pixel intensity. Representative fluorescence microscopy image of beads (inset). (c) cDNA copies of reovirus RNA relative to Gapdh as a function of the number of custom primers included in the ligation reaction (bulk assay). (d) Enrichment of PCR amplicons relative to Gapdh in DART-seq versus Drop-seq libraries as a function of distance to the target locus. (Mridusmita S, et al. 2018)
Figure 1. Characterization of DART-seq primer bead synthesis and comparison of DART-seq and Drop-seq. (a) Number of fluorescence probes bound per bead as a function of the number of primers per bead included in the ligation reaction (four distinct primer sequences). Schematic of fluorescence hybridization assay (inset), (b) Bead-to-bead variability in fluorescence pixel intensity. Representative fluorescence microscopy image of beads (inset). (c) cDNA copies of reovirus RNA relative to Gapdh as a function of the number of custom primers included in the ligation reaction (bulk assay). (d) Enrichment of PCR amplicons relative to Gapdh in DART-seq versus Drop-seq libraries as a function of distance to the target locus. (Mridusmita S, et al. 2018)
We offers complete, professional droplet-assisted RNA targeting by single-cell sequencing service, as well as customized experimental protocols based on your project requirements and sample characteristics.
The Drop-seq technology encapsulates individual cells in microbeads with barcodes, and initiates the process of reverse transcription of mRNA of the cells. Before performing routine Drop-seq analysis, our R&D team designed an effective method to customize microbeads in order to recover and analyze more types of RNA molecules and conduct a more comprehensive analysis of single cells. At the same time, the technology can identify cells infected by the virus, quantify the expression of virus and host genes, and then detect the host's response to infection at the single-cell level.
01
Personalized experimental plan
02
Experienced expert team
03
Innovative technical approach
04
Complete and detailed project report

We can customize services and adjust and optimize specific experimental plans according to your needs so that they can better fit your project conditions. We provide comprehensive and reliable services and can submit detailed project analysis reports to you.
For more information or to discuss in detail, please contact us.
Lifeasible provides comprehensive plant breeding solutions and a diverse technology platform to meet the plant breeding research needs of global customers. In addition, we provide plant genetic engineering services to support breeding research.
Reference
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024
Application of Exosomes in Facial Beauty
April 12, 2024