Rice, one of the world's most important staple crops, is primarily affected by various diseases, including bacterial blight, rice blast, and bacterial leaf streak, resulting in significant economic losses. A deeper understanding of the molecular mechanisms underlying pathogen infection and the factors that determine host susceptibility or resistance are of great scientific and practical significance for improving crop resilience, stabilizing agricultural production, and ensuring food security.
In recent years, research on plant long noncoding RNAs (lncRNAs) has rapidly advanced, revealing their important roles in regulating growth and development, environmental adaptation, epigenetic inheritance, and stress resistance. However, there are currently few reports on the molecular functions of susceptibility lncRNAs (S lncRNAs) as novel targets for regulating broad-spectrum crop resistance without affecting growth.
On August 18, 2025, a team led by Yu-chan Zhang from Sun Yat-Sen University published a research paper titled "Gene Editing of a Susceptibility LncRNA Enhances Broad-Spectrum Disease Resistance in Rice without Developmental Trade-Offs" in Advanced Science.
The study revealed that upon pathogen invasion, the long noncoding RNA RESIS is activated by effector-binding elements on its promoter. It then binds to two core components of the NatA complex, NAA15 and NAA10, and enhances the activity of the NatA complex in protein N-terminal acetylation, promoting pathogen infection. Knockout of RESIS inhibits this process and enhances translation during pathogen invasion, conferring resistance to fungal and bacterial diseases without the growth inhibition typically associated with direct knockdown/deletion of the NatA complex. These findings highlight the potential of S lncRNAs as targets for enhancing broad-spectrum disease resistance in crops, particularly in balancing resistance and growth, offering great promise for practical applications.
By analyzing transcriptome data from japonica, indica, and common wild rice, and combining it with transcriptome data from bacterial blight infection, the research team discovered a highly conserved lncRNA that is significantly upregulated upon bacterial blight infection. A T-DNA insertion-inactivated mutant of this lncRNA exhibited enhanced resistance to bacterial blight, and thus the lncRNA was named RESIS (REsponsIve to Xoo and conServed). Further bacterial blight infection experiments with the constructed RESIS overexpression (RESIS-OE) and knockout mutants (resis) revealed that the RESIS-OE strain was significantly susceptible, while the resis strain exhibited significant resistance, suggesting that RESIS facilitates the pathogen's infection process in rice. Observation and statistical analysis of agronomic traits in the mutant plants revealed that knockout of RESIS did not affect plant yield, 1000-grain weight, or plant architecture, and that knockout of RESIS rescued the significant yield reduction caused by bacterial blight infection. The resis mutant also exhibited good resistance to bacterial leaf streak and blast, highlighting the lncRNA RESIS as a promising susceptibility target for achieving broad-spectrum resistance in rice without compromising other agronomic traits.
Using RNA pulldown and RNA TurboID experiments, the research team identified NAA15 and NAA10, two proteins specifically binding to RESIS. RIP and TriFC experiments further validated the interaction between RESIS and these proteins.
NAA15 and NAA10 are core members of the NatA complex, a key N-terminal acetylation complex, accounting for over 40% of the proteomes of higher animals and plants. NatA-mediated N-terminal acetylation plays a crucial role in regulating rice growth and development and responses to abiotic stresses. The NatA complex is crucial for plant growth and development and stress responses. Consistent with previous studies, homozygous knockout plants for NAA15 and NAA10 were unavailable. Phenotypic observations of NAA15-RNAi and NAA10-RNAi plants revealed significant growth inhibition, affecting plant height and panicle length. Compared to control plants, panicle branch number, grain number, and yield per plant were significantly reduced, indicating that NAA15 and NAA10 play a key role in maintaining rice growth, development, and yield.
Resistance to bacterial blight in transgenic plants was observed and statistically analyzed. After treatment with PXO99A, NAA15-RNAi and NAA10-RNAi plants exhibited significant resistance to bacterial blight. Further experiments with NAA15-RNAi and NAA10-RNAi plants against rice blast and bacterial leaf streak revealed significantly enhanced resistance to both diseases.
These results suggest that NAA15 and NAA10 function similarly to RESIS during pathogen infection. RESIS may regulate plant resistance to pathogens by affecting both.
Further investigation into the regulatory relationship between RESIS and NAA15 and NAA10 revealed that RESIS evolved a specific domain during domestication that directs NAA10 binding and catalyzes N-terminal acetylation of proteins involved in ribosome assembly and translation, thereby playing a role in disease resistance. Because knocking down NAA15 or NAA10 enhances resistance, these genes alone are unsuitable targets for enhancing crop disease resistance. However, targeting RESIS avoids these disadvantages and has significant potential for practical applications.
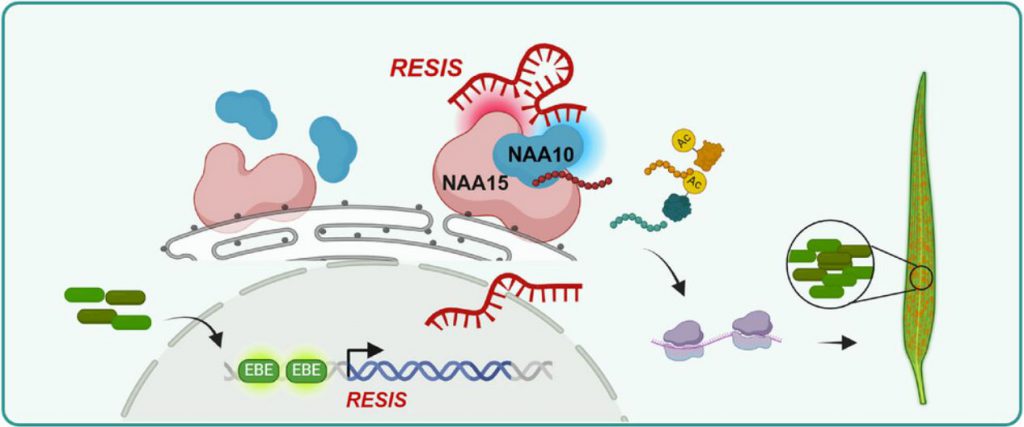
Figure 1. Working model of the RESIS module: Upon pathogen invasion, RESIS is activated through the EBEs on its promoter region. (Zhao, et al. 2025)
In summary, this study proposes a novel strategy for enhancing broad-spectrum crop resistance. By editing S lncRNA RESIS, broad-spectrum crop resistance to pathogens can be enhanced without compromising growth, development, or yield. The significant potential of S lncRNAs in crop resistance research provides a new direction for future crop genome editing.