Gene editing technology offers unlimited possibilities for crop genetic improvement and functional genomics research. However, two major technical bottlenecks have long hindered the full realization of its potential. First, the reliance on time-consuming, labor-intensive, and species-dependent tissue culture regeneration systems. Second, after obtaining edited progeny, tedious selfing or hybridization is required to isolate the exogenous transgenic elements to meet regulatory requirements for non-GMO crops.
It can be said that whoever can first overcome these two constraints will hold the "golden key" to future plant biotechnology competition.
On September 19, 2025, Degao Liu's team from Texas Tech University published a new paper titled "Heritable, Tissue Culture-Independent and Transgene-Free Genome Editing in Plants via Viral Delivery of CRISPR/AsCas12f" in the Plant Biotechnology Journal. This study reports a novel plant gene editing strategy, using an RNA viral vector to deliver the CRISPR/AsCas12f system into plants. They successfully achieved heritable, tissue culture-independent, and transgene-free "one-step" gene editing in Nicotiana benthamiana and tomato plants, providing an efficient solution to address the two aforementioned bottlenecks.
RNA viral vectors are ideal delivery vehicles for transgene-free transient expression and systemic delivery. However, viral vectors have limited loading capacity and cannot accommodate SpCas9. Therefore, this study selected AsCas12f, which consists of only 422 amino acids.
The research team cloned an AsCas12f variant and its sgRNA components into an expression vector derived from tobacco rattle virus (TRV). TRV, an RNA virus, can replicate and move long distances within plants, delivering editing tools to various sites, including the shoot apical meristem, without integrating its own genetic material into the plant genome, thus paving the way for transgene-free editing.
The research team first conducted proof-of-concept testing in tobacco. They designed a sgRNA targeting the PDS gene and introduced the recombinant TRV virus into tobacco plants via Agrobacterium-mediated leaf injection.
Approximately two weeks after inoculation, albinism was observed on new tobacco leaves. As the plants grew, the albinism gradually spread to the stems and calyxes. Next-generation sequencing of these leaves revealed an 83.08% frequency of indels at the target site, with multinucleotide deletions predominantly occurring.
More importantly, whether this editing effect could be transmitted to the next generation was crucial. The researchers harvested seeds from virus-infected plants at different stages of development. Among the germinated T1-generation seedlings, pure albinism was observed at a frequency of 5.67%. Sequencing confirmed that all albinism seedlings were homozygous or biallelic mutants in the NbPDS-1 gene. Even in phenotypically normal green T1-generation seedlings, extremely high gene editing frequencies were detected, with some batches achieving 100% editing rates. PCR testing confirmed that no traces of the TRV viral vector were present in any of the edited progeny.
These results clearly demonstrate that the TRV-AsCas12f system successfully achieved efficient, heritable, tissue culture-free, and transgenic gene editing in N. benthamiana.
The research team further applied this strategy to tomatoes, targeting the SIAN2 gene, which controls anthocyanin synthesis. The mutant stems changed from purple to green. The authors injected the virus directly into the axillary meristem of tomatoes, hoping to directly edit the stem cells that develop into lateral branches and inflorescences.
However, the tomato's robust antiviral defense system inhibited viral replication and movement at normal temperatures, resulting in initial failure. To address this, the research team employed two ingenious strategies:
Both strategies were successful, resulting in multiple SIAN2-edited lateral branches that exhibited the desired green phenotype. The low-temperature treatment strategy was particularly effective, achieving a total editing efficiency of 42.11%. Likewise, these edits are stably inherited to the next generation, and subsequent plants are free of viral transgenes.
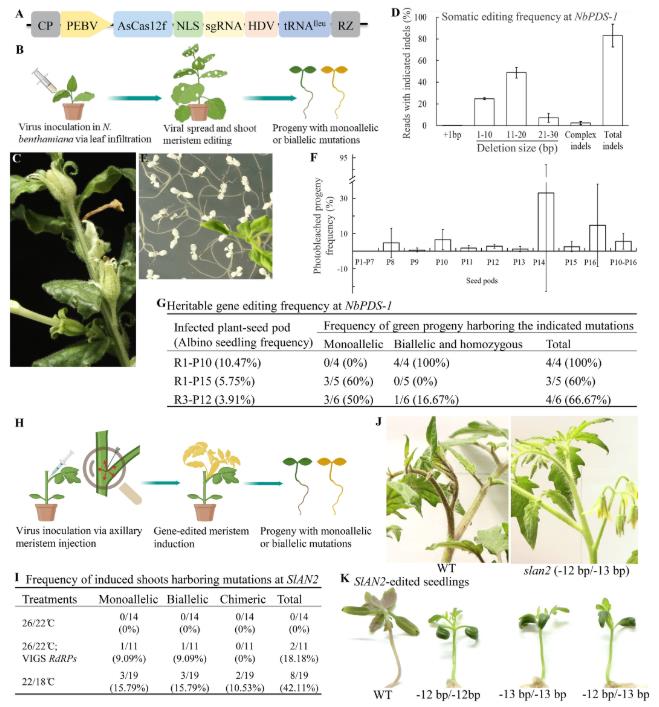
Figure 1. Heritable, tissue culture-independent, and transgene-free genome editing in Nicotiana benthamiana and tomato via viral delivery of CRISPR/AsCas12f. (Hu, et al. 2025)
This study combined the miniaturized CRISPR/AsCas12f system with RNA viral delivery, supplemented by strategies to overcome host antiviral defenses. This study successfully established a novel gene editing process in model plants and important economic crops that is heritable, tissue culture-free, and transgenic-free.
This technology is not only simple to use and has a short turnaround time, but also produces edited crops that are free of foreign DNA, effectively circumventing regulatory barriers for genetically modified crops. This one-stop solution opens a new path for accelerating basic plant science research and precision genetic improvement of crops.