On October 21, 2025, a team led by Hongjie Li from Xianghu Laboratory and his collaborators published a research paper entitled "Wheat powdery mildew resistance gene Pm52/Pm6 from Triticum timopheevii encodes an NLR integrated with PRK domain" in Plant Communications.
This study successfully cloned the powdery mildew resistance gene Pm52 from the main wheat varieties Liangxing 99 and Jimai 22 using map-based cloning, PacBio HiFi, and Hi-C sequencing technologies. This gene, derived from T. timopheevii, encodes a novel NLR-PRK fusion protein with broad-spectrum resistance, and it was confirmed that Pm52 and Pm6 are the same gene. Simultaneously, the functional molecular marker Pm52-FM was developed, providing a reliable tool for marker-assisted selection in wheat breeding and facilitating the rapid and efficient application of this resistance gene.
Wheat powdery mildew is an epidemic fungal disease that causes severe losses to wheat production. Although several powdery mildew resistance genes have been cloned, their breeding applications are limited by issues such as linkage carryover. Discovering resistance genes from widely cultivated high-yielding varieties has become a key breakthrough in breeding high-yielding, stable-yielding, and disease-resistant wheat varieties. Integrating these genes into superior genetic backgrounds can effectively avoid linkage carryover and can be directly used for wheat powdery mildew resistance breeding.
Liangxing 99 and Jimai 22 are major winter wheat varieties with high powdery mildew resistance. Jimai 22 has ranked first in planting area in China for 11 consecutive years, with a cumulative planting area of over 400 million mu (approximately 66.7 million hectares) for both varieties. The powdery mildew resistance gene MlLX99 in Liangxing 99 is located in an 8.4 cM genetic region on chromosome 2BL between markers BE604758 and Xgwm120, and was later officially named Pm52. The powdery mildew resistance gene of Jimai 22 was also located on chromosome 2BL, the same Pm52 gene as Liangxing 99.
Using the functional markers TaPm2-F1/TaPm2-R1 for Pm2, it was found that both Liangxing 99 and Jimai 22 carry the Pm2 gene. To eliminate interference from Pm2 on Pm52 gene cloning, 12 wheat powdery mildew strains were analyzed, and a specific strain B37 capable of distinguishing between Pm2 and Pm52 resistance was screened. Pm52 showed a resistant response to B37, while Pm2 showed a susceptible response.
Genetic analysis using a recombinant inbred line population of Liangxing 99/Zhongzuo 9504 confirmed that the resistance of Liangxing 99 to strain B37 is controlled by a single gene. Using the Chinese Spring genome as a reference, molecular markers were developed, and Pm52 was located within a 0.18 cM genetic interval between markers Wggbq10 and Wggbq6. Further, using a large-scale F2 population, Pm52 was finely mapped to the region between markers Wggbq44 and WggbH612-5, corresponding to the physical region of chromosome 2B of the Chinese spring reference genome, spanning 694.50–718.80 Mb. All eight polymorphic markers developed within this 24.3 Mb region co-segregated with the Pm52 gene, indicating significant recombination repression in this region, making it difficult to pinpoint the target gene using traditional chromosome walking.
To overcome recombination repression and clone the Pm52 gene, PacBio HiFi and Hi-C sequencing and genome assembly were performed on Jimai 22. Comparative genomic analysis revealed significant structural variations within the Pm52 mapping region. The flanking markers Wggbq44 and WggbH612-5 correspond to a 14.83 Mb region in the Jimai 22 genome, annotating a total of 279 genes, including 8 NLR genes and 24 kinase genes.
Through EMS mutagenesis, 11 susceptible mutants of Liangxing 99 were obtained. Candidate genes were amplified and sequenced using gene-specific primers. All mutants were found to be mutated in the TraesJM222B679606 gene, including 13 missense mutations and 2 premature termination mutations. This gene mutation resulted in loss of powdery mildew resistance, indicating that TraesJM222B679606 is a candidate gene for Pm52.
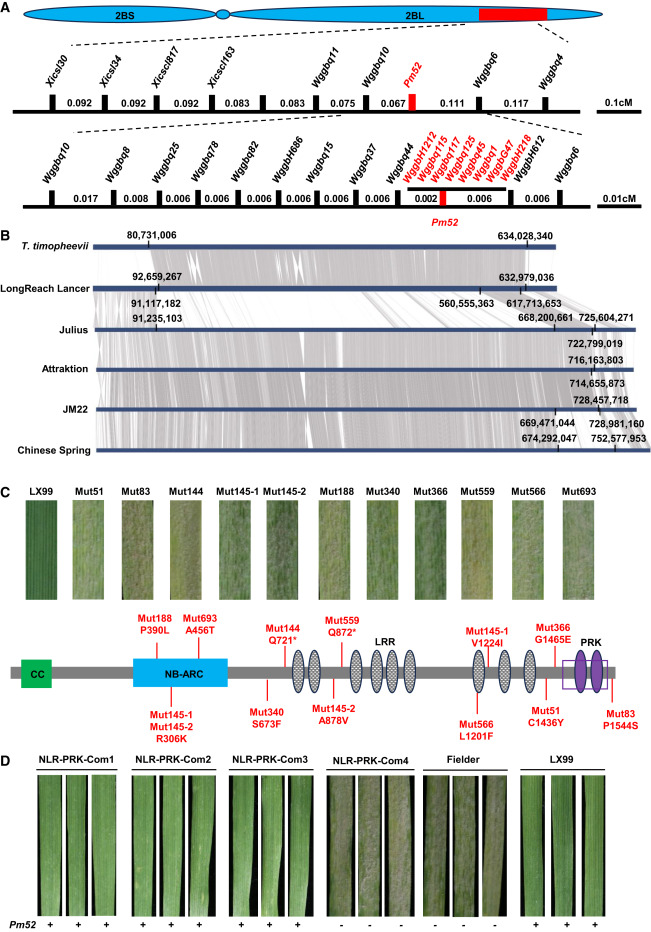
Figure 1. Map-based cloning and functional verification of the Pm52 gene. (Qiu, et al. 2025)
TraesJM222B679606 encodes an NLR protein containing complete CC, NBS, and LRR domains, with a PRK domain fused to its C-terminus; therefore, this gene was named NLR-PRK. Its full-length cDNA was obtained using 5' and 3' RACE techniques, revealing that its coding region contains only one exon. The 3'-UTR generates six transcriptomorphs (TV1-TV6) through alternative splicing, with TV1 being the predominant form. qRT-PCR analysis showed that all six transcripts were significantly upregulated 24 and 48 hours after powdery mildew inoculation.
To verify the disease resistance function of NLR-PRK, a transgenic vector ProNLR-PRK:NLR-PRK (total length 11,731 bp, containing a 4,731 bp coding region, a 3,000 bp upstream regulatory region, and a 4,000 bp downstream regulatory region) was constructed, driven by its own promoter. Through Agrobacterium-mediated genetic transformation, the vector was introduced into the susceptible wheat variety Fielder, resulting in three transgenic lines. T1 generation inoculation identification showed that all NLR-PRK positive transgenic plants exhibited high resistance to powdery mildew, while negative plants and Fielder showed high susceptibility, confirming that NLR-PRK is the Pm52 gene.
Resistance identification using eight powdery mildew strains revealed that the Pm52 transgenic lines showed resistance to six strains consistent with Liangxing 99 (Pm2+Pm52) and Jimai 22 (Pm2+Pm52), while Nongda 399 (Pm2) showed resistance to only two strains. This result further confirms the disease resistance function of Pm52.
Sequence alignment showed that the Pm52 genome sequence differs significantly from most published wheat genomes, but is completely identical to the sequences of LongReach Lancer, Julius, Attraktion, and T. timopheevii. Chromosome 2B of LongReach Lancer contains a 472 Mb fragment of T. timopheevii. Genome alignment revealed that Attraktion, Julius, LongReach Lancer, and Jimai 22 all carry the wheat-T. timopheevii translocation chromosome T2B·2G, proving that the segment containing Pm52 originates from T. timopheevii. Further analysis showed that Jimai 22 has the highest sequence similarity to Attraktion and Julius, while LongReach Lancer has the highest sequence similarity to T. timopheevii. It is speculated that the T2B·2G translocation fragments of Jimai 22, Attraktion, and Julius may originate from LongReach Lancer.
The functional marker Pm52-FM, developed using the NLR-PRK gene sequence, can amplify a Pm52-specific band in the varieties Coker 747 and Coker 783, which carry the Pm6 gene (derived from T. timopheevii). The full-length Pm52 sequence of these two varieties is completely identical to that of Liangxing 99 and Jimai 22, confirming that Pm52 and Pm6 are the same gene.
Analysis of 1368 germplasm resources using Pm52-FM revealed the Pm52 gene in only 3 T. timopheevii accessions and 5 common wheat materials from the former Soviet Union, Portugal, Spain, and Tunisia. This provides evidence of the gene's infiltration into Chinese varieties via Eurasian and North African germplasm.
Liangxing 99 and Jimai 22 are not only widely planted as superior varieties in production but have also been used as parents in the breeding of numerous new varieties. Analysis of 1613 Chinese wheat varieties and breeding lines using Pm52-FM identified 85 materials containing Pm52, all of which are highly resistant to powdery mildew strain B37. Cloning of the Pm52 gene and the development of the functional marker Pm52-FM provide a reliable tool for marker-assisted selection breeding of wheat, significantly accelerating the efficient application of the Pm52 gene in wheat disease resistance breeding.