Epigenetics, as molecular markers that do not alter DNA sequence, can influence various aspects of eukaryotic cell growth and development, as well as adaptation to environmental changes. Theoretically, stable and heritable epigenetic variation is beneficial to improving plant adaptability. However, establishing a link between plant population genetics and potential epigenetic polymorphisms currently faces several limitations. DNA methylation, as a heritable epigenetic molecular marker, can occur in both transposon regions (transposable element methylation, teM) and genobody regions (gene-body methylation, gbM) of genes. teM mainly occurs at CG, CHG, and CHH (H:A,T, or C) sites and is usually negatively correlated with gene expression. gbM, on the other hand, only occurs at CG sites and is related to the stability and precision of gene expression within plant species, potentially helping to suppress the production of abnormal transcripts. However, prior to this study, there was no direct evidence to clearly demonstrate the causal regulatory role of gbM on gene expression in natural populations or its contribution to complex traits.
On September 12, 2025, Daniel Zilberman's team from the John Innes Centre published a significant research finding in Nature Plants entitled "Gene body methylation regulates gene expression and mediates phenotypic diversity in natural Arabidopsis populations."
This study is the first to systematically reveal in natural Arabidopsis populations that gbM, independent of genetic variation, significantly regulates gene expression and drives multiple phenotypic variations, including flowering time, stress resistance, and mineral accumulation. Through large-scale epigenome-wide association (epiGWA), the team precisely located several new genes affecting adaptive traits and discovered that gbM variations are significantly associated with environmental factors such as air pollution (e.g., NO2), providing a new epigenetic perspective for understanding the rapid adaptation of plants to climate change. This research not only confirms the important role of epigenetic variation in natural evolution but also provides new strategies for future crop breeding and ecosystem conservation.
Studies have found that gbM and teM are distributed differently and independently in natural Arabidopsis populations. gbM is prevalent in constitutively high-expression genes, while teM is more common in low-expression or tissue-specific genes. Through mathematical modeling of gbM dynamics, researchers found that the steady-state distribution of gbM can be accurately predicted without introducing teM, further supporting their independence.
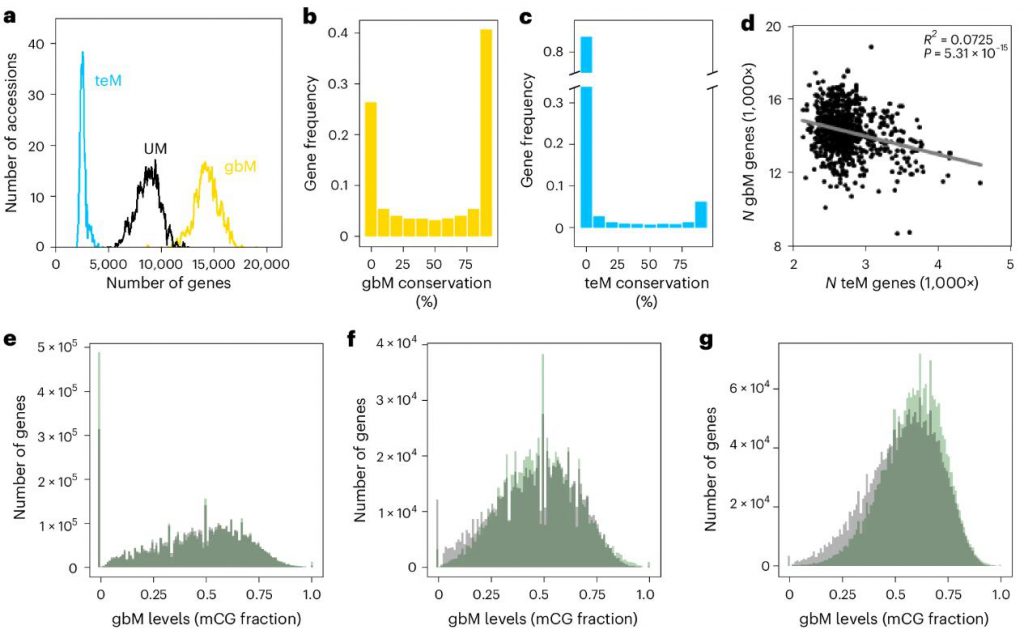
Figure 1. GbM and teM are independent phenomena. (Shahzad, et al. 2025)
Across the entire genome, gbM and teM explain 15.2% and 26.0% of gene expression variation, respectively, comparable to the contribution of SNPs (23.5%). gbM has a greater impact on the expression of highly conserved genes, while teM has a more significant impact on low-entropy (tissue-specific) genes, indicating that they have complementary roles in different gene networks.
Analysis of transcriptome data from the met1 mutant (gbM deletion) revealed that gbM loss significantly affects the expression of its associated genes: positively correlated eQTL gbM gene expression decreased, while negatively correlated eQTL gbM gene expression increased. This effect was dose-dependent with gbM levels and consistent across multiple tissues and genetic backgrounds, confirming that gbM directly regulates gene expression.
Through epiGWA, researchers identified several gbM loci significantly associated with adaptive traits such as flowering time, heat and drought tolerance, and mineral accumulation. Mutant validation showed that nearly 90% of candidate genes indeed regulate the corresponding phenotypes, and epiGWA is more precise in locating causal genes than traditional GWA because it overcomes the limitations of linkage disequilibrium.
The study found that gbM variation is significantly associated with 171 environmental variables, particularly with factors such as temperature, precipitation, and atmospheric NO2. For example, the gbM status of the FLC gene was significantly correlated with spring NO2 concentration, and the NO2 concentration was highly consistent with flowering time in the laboratory, suggesting that gbM may help Arabidopsis thaliana adapt rapidly to anthropogenic environmental changes.