Epigenetic molecular mechanisms, including methylation of cytosines in DNA, covalent modifications of histones, and RNA interference (RNAi) pathways, are involved in the regulation of pivotal cellular processes such as gene expression, replication timing, differentiation, and adaptation of the organism to changing environmental conditions. As sessile organisms, plants have developed a complex set of epigenetic tools to cope with suboptimal living conditions and adapt to environmental changes.
 Fig. 1 Brief diagram of epigenetic modifications of plant telomeres.
Fig. 1 Brief diagram of epigenetic modifications of plant telomeres.
Epigenetic regulators have pivotal roles in controlling the chromatin state at telomeres and sub-telomeric regions, contributing to maintaining chromosomal homeostasis in plants. At Lifeasible, our team of highly skilled scientists and researchers possess extensive knowledge in telomere biology and epigenetic modifications.
Histone Modifications
- In eukaryotes, the combination of different post-translational modifications (methylation, acetylation, phosphorylation, ubiquitination, SUMOylation, and ADP-ribosylation) on the N-terminal tails of histone dictates the rapid change of the chromatin state into a transcriptionally-active euchromatin or silent heterochromatin state.
- Lifeasible offers cutting-edge technologies for analyzing histone modifications at plant telomeres. Methylation of histone H3 at lysine 9 (H3K9me) and histone H4 at lysine 20 (H4K20me) are associated with telomere silencing. These repressive marks recruit chromodomain-containing proteins, such as HP1 (heterochromatin protein 1), leading to a condensed heterochromatic state at telomeres. On the other hand, acetylation of histones, such as H3K9ac and H4K16ac, is linked to telomere elongation and increased telomerase activity.
- Our advanced techniques allow for precise quantification and characterization of histone marks, enabling researchers to unravel the intricate regulatory network governing telomere function, including but not limited to yeast two-hybrid screening, Southern blot analysis of mutants, telomere ChIP, and more.
DNA Methylation
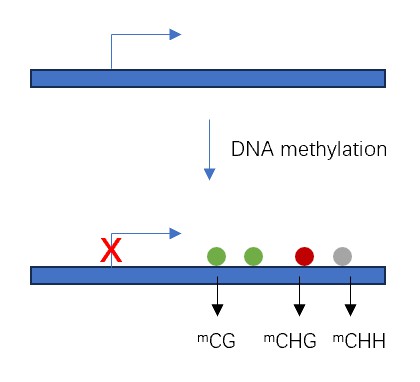
- We also offer an analysis of DNA methylation, an essential epigenetic mechanism involved in telomere regulation. DNA methylation is adding a methyl group to the carbon 5 position of cytosine residues, predominantly occurring in a CpG dinucleotide context.
- We help our clients understand the regulatory role of DNA methylation in plant telomeres. Hypermethylation of sub-telomeric regions is generally associated with telomere shortening, while hypomethylation is associated with telomere elongation. Furthermore, DNA methylation can also modulate telomeric chromatin structure and telomere position effect.
- Our state-of-the-art methods enable precise quantification of DNA methylation levels and identification of differentially methylated regions (DMRs) at telomeres, facilitating the elucidation of the intricate relationship between telomere function and DNA methylation patterns.
With Lifeasible as your trusted partner, you can be confident in pursuing groundbreaking discoveries in the realm of epigenetic modifications of plant telomeres. If you are interested in our services or have some questions, please feel free to contact us or make an online inquiry.
Reference
- Nisa MU, et al. (2019). "The Plant DNA Damage Response: Signaling Pathways Leading to Growth Inhibition and Putative Role in Response to Stress Conditions." Front Plant Sci. 10, 653.
Our products/services are For Research Use Only. Not For Clinical Use!
 Fig. 1 Brief diagram of epigenetic modifications of plant telomeres.
Fig. 1 Brief diagram of epigenetic modifications of plant telomeres.