On September 23, 2025, a team led by Fanny Bellegarde from Nagoya University published a research paper in Plant Communications entitled "Fluctuation in nitrate availability impacts cytokinin biosynthesis through histone modifications of IPT3 in Arabidopsis roots for growth acclimation." This study systematically elucidates the molecular mechanism by which changes in nitrate supply precisely control the transcription of IPT3, a key gene in cytokinin synthesis, through dynamic regulation of histone modifications H3K4me3 and H3K27me3, thereby regulating the root-shoot-end balance of plants to adapt to nutrient fluctuations.
Nitrate is one of the most important nitrogen sources for plants, but its availability in natural soils often fluctuates dramatically due to factors such as rainfall and microbial activity. To cope with this instability, plants have evolved mechanisms to optimize growth by sensing and rapidly adapting to changes in nutrient availability. Cytokinin, as an important plant hormone, plays a central role in the transmission of nitrate signals from roots to aboveground parts and in coordinating overall growth. IPT3 is a key gene in nitrate-induced cytokinin biosynthesis. However, the epigenetic regulatory mechanism by which plants rapidly and precisely regulate IPT3 expression to adapt to dynamically changing nitrate environments remains unclear.
Time-series experiments revealed that changes in nitrate supply rapidly alter the dynamic levels of the IPT3 gene expression in Arabidopsis roots, accompanied by significant fluctuations in cytokinin levels. When nitrate was deprived, IPT3 expression was inhibited within 2 hours, and the levels of active cytokinins (such as iP and tZ) decreased to basal levels within 24 hours. Upon restoration of nitrate supply, IPT3 expression was rapidly induced within 15 minutes, exhibiting a two-phase upward trend.
Further research revealed that this rapid transcriptional regulation of the IPT3 gene is closely related to the state of histone modifications in its chromatin. Under nitrate-sufficient conditions, the IPT3 gene region is enriched with the activation marker H3K4me3. When subjected to nitrate starvation, the repressive marker H3K27me3 gradually deposits, while H3K4me3 levels decrease, both contributing to gene silencing. Crucially, the rapid induction phase following nitrate resupply is accompanied by the rapid removal of H3K27me3, while subsequent sustained high expression depends on the redeposition of H3K4me3.
Using a series of histone-modifying enzyme mutants, the research team identified key chromatin effectors involved in this process. The study found that the H3K4me3 demethylase JMJ14 mediates transcriptional repression of IPT3 in the early stages of starvation; while the H3K27me3 methyltransferase CLF is responsible for maintaining the stable silencing state of the gene after prolonged starvation. During the nitrate resupply induction phase, H3K27me3 demethylation is mainly performed by REF6/ELF6/JMJ13, while H3K4me3 deposition depends on the methyltransferase ATX1. Furthermore, while the major nitrate-responsive transcription factors NLP6/NLP7 are essential for the full induction of IPT3, the removal of H3K27me3 does not depend on them. Therefore, the early stages of nitrate supplementation-induced IPT3 require the removal of H3K27me3 by demethylases such as REF6, ELF6, and/or JMJ13 to reactivate this site, followed by stimulation by NLP6/NLP7. The further significant increase in IPT3 transcription levels 3 hours after nitrate supplementation depends on this process mediated by ATX1 through the deposition of H3K4me3.
Studies have shown that the dynamic changes in these histone modifications directly affect the biosynthetic output of root cytokinins. In the corresponding mutants, alterations in IPT3 transcription levels led to abnormal levels of iP and tZ cytokinins, thereby affecting the plant's growth phenotype under fluctuating nitrate conditions, particularly root growth rate and root-to-shoot ratio. Therefore, fine-tuning IPT3 expression through histone modifications is a crucial step in coordinating the allocation of growth resources among plant organs and achieving rapid growth adaptation.
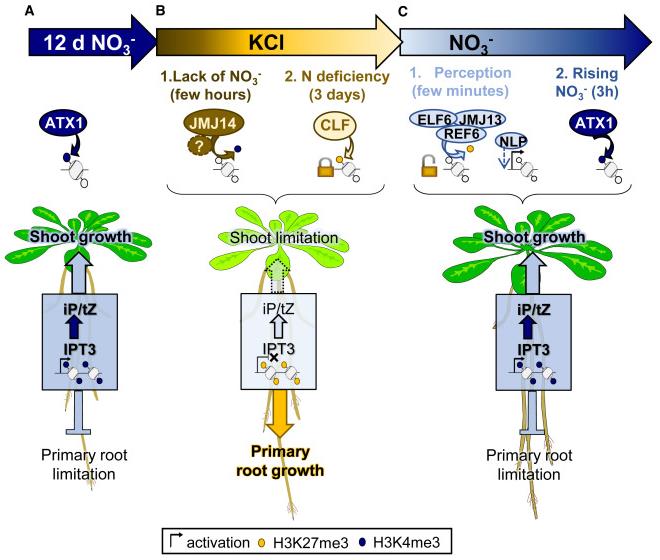
Figure 1. Model of chromatin-mediated CK biosynthesis regulation during fluctuating nitrate and its impact on growth. (Bellegarde, et al. 2025)
This study systematically elucidates the molecular pathway by which Arabidopsis thaliana dynamically regulates cytokinin biosynthesis through epigenetic mechanisms to adapt to fluctuations in nitrate levels. The study confirms that the transcription of the key synthetic gene IPT3 is precisely and dynamically regulated by histone modifications H3K4me3 and H3K27me3. In the early stages of nitrate starvation, JMJ14-mediated H3K4me3 demethylation synergistically inhibits IPT3 expression with an unknown repressor protein; prolonged starvation stabilizes its silenced state through CLF-mediated H3K27me3 deposition. Conversely, after nitrate resupply, REF6/ELF6/JMJ13-catalyzed H3K27me3 demethylation is a prerequisite for gene "unlocking" and early induction, while ATX1-mediated H3K4me3 deposition drives sustained high expression in the later stages. This temporal change in chromatin state directly determines the synthesis and output of root cytokinins, thereby regulating root-cap growth balance and achieving rapid growth adaptation. This discovery not only reveals a new epigenetic level of the cross-interaction between nutrient signals and hormonal regulation, but also provides important theoretical basis and molecular targets for future research on improving crop nutrient use efficiency by manipulating chromatin state.
This research provides an integrative perspective on plant adaptation to nitrate fluctuations, including its temporal progression and molecular mechanisms. The research team identified several chromatin effector factors worthy of in-depth analysis, particularly laying the foundation for studying their interactions with transcription factors. This study reveals another regulatory interface between nitrate nutrition and hormonal signals that regulate plant growth, opening up new research areas for understanding the complex interactions between chromatin modification, hormones, and nutrient regulation to elucidate the mechanisms by which plants adapt to fluctuating nutrient environments. This understanding is expected to provide possibilities for developing new plant breeding strategies in the future.