On October 23, 2025, a research team led by Umar F. Shahul Hameed and Stefan T. Arold from King Abdullah University of Science and Technology (KAUST) published a research paper entitled "Cryo-EM structural analyses reveal plant-specific adaptations of the CDC48 unfoldase" in Plant Communications. This study, using cryo-electron microscopy and biochemical analysis, discovered that Arabidopsis thaliana AtCDC48A can independently bind the cofactors UFD1 and NPL4, and that only NPL4 is required to mediate substrate unfolding.
CDC48 is an AAA+ ATPase widely involved in protein degradation, quality control, and membrane fusion. It recognizes ubiquitinated substrates and uses energy provided by ATP hydrolysis to extract or unfold them from the complex, subsequently delivering them to the proteasome for degradation. This process is crucial for maintaining proteomic stability and enabling rapid environmental adaptation in organisms. The CDC48 system in animals and fungi has shown structural and functional conservation, but research on its plant homologs remains almost nonexistent.
Protein domain analysis shows that the domain composition and sequence of CDC48 are highly conserved in higher plants, lower plants, and even eukaryotes. To explore plant-specific adaptive mechanisms, researchers used single-particle cryo-electron microscopy to resolve the structure of the recombinant full-length AtCDC48A from A. thaliana. AtCDC48A from A. thaliana exhibits high conservation with its animal and fungal homologs in core structural features such as the active site and conformational changes in the N-terminal domain (NTD) induced by nucleotide binding. However, the study also revealed two key plant-specific differences. First, when binding to the ATP analogue (AMPPNP), AtCDC48A clearly exhibits an "NTD-down" conformation, which is extremely rare in its homologs. Second, no large-scale rotation of the D2 domain relative to D1 occurs during the catalytic cycle. Furthermore, the function of its C-terminal extension changes with the nucleotide state, and it primarily exhibits a monohexamer state in solution, rather than the D2-mediated dodecamer commonly seen in animal models.
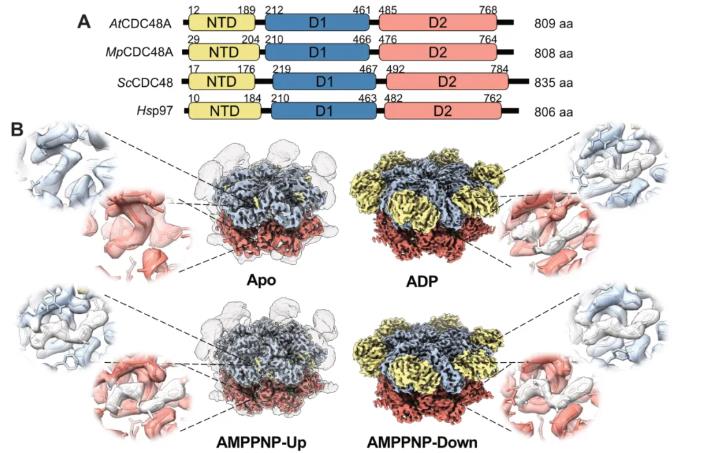
Figure 1. Idiosyncrasies of AtCDC48A. (Huntington, et al. 2025)
Functional experiments showed that the AtCDC48A cofactors AtUFD1 and AtNPL4 do not form stable heterodimers, but can bind to AtCDC48A independently. AtNPL4 alone can mediate the unfolding of ubiquitinated substrates, while the presence of AtUFD1 significantly enhances the unfolding efficiency, demonstrating a synergistic effect between the two. This pattern contrasts sharply with the mechanism in animals and yeast where a heterodimer must be formed.
Further structural analysis and mutation experiments revealed that plant NPL4 lacks the zinc finger domain (ZnF) commonly found in animals, but it establishes a stable interaction with CDC48 through its unique UBXL–MPN linker, compensating for the loss of binding force due to ZnF deficiency. This plant-specific binding mechanism not only enhances the interaction between NPL4 and CDC48 but may also play a crucial role in regulating substrate recognition and initiating unfolding.
Evolutionary analysis of multiple eukaryotic groups revealed profound evolutionary significance in the modular, independent binding pattern of CDC48 with UFD1 and NPL4 in plants. While the CDC48-UFD1 interaction is conserved across all eukaryotes, the zinc finger domain of NPL4 and its forced heterodimer formation with UFD1 are only observed in ostracods. In contrast, plants and other basal eukaryotic groups retain mechanisms for direct anchoring of CDC48 through simple structural motifs.
This study, through high-resolution structural analysis and functional experiments, systematically revealed the structural and functional characteristics of the plant CDC48 system, discovering significant differences from animals and yeast in its cofactor binding mechanisms, structural dynamics, and evolutionary pathways. These differences reflect the unique adaptations of plants in protein degradation mechanisms and provide an important foundation for future research in plant molecular biology and agricultural biotechnology.