On November 7, 2025, a team led by Jianbing Yan from Huazhong Agricultural University and their collaborators published a research paper titled "Genetic basis of flavor complexity in sweet corn" in Nature Genetics, systematically revealing the genetic basis of sweet corn flavor formation. This research not only breaks the traditional breeding concept that "sweet equals delicious," but also proposes a new direction for "flavor design" breeding, providing a scientific basis and technical path for cultivating high-yield and delicious sweet corn varieties and achieving high-quality and high-efficiency upgrading of the sweet corn industry.
As a widely cultivated and important economic crop globally, sweet corn is not only a nutritious food source but also an important raw material for the food industry. With consumers' increasing demand for high-quality agricultural products, the flavor of sweet corn is no longer limited to the simple dimension of "sweetness," but rather a comprehensive reflection of multiple sensory experiences, including sweetness, mouthfeel, aroma, and texture. Traditional breeding methods mainly rely on phenotypic selection and empirical judgment, and have not yet systematically analyzed and utilized the genetic basis of flavor, which undoubtedly limits the efficiency of breeding high-quality varieties. Therefore, constructing a flavor resource and knowledge system based on genetic information has become an important research direction in the interdisciplinary field of sweet corn genetic improvement and even food science.
The research team systematically analyzed the genetic basis of sweet corn flavor by using multi-omics analysis on 295 sweet corn germplasm resources. The study shows that super sweet corn originated from a transposon-induced inversion in the Sh2 gene, while ordinary sweet corn originated from a spontaneous point mutation in the Su1 gene. Besides these two significant sites, there is also significant genetic differentiation between sweet corn and ordinary field corn at the whole-genome level. Sweet corn has undergone strong positive selection during evolution, especially in genes related to carbohydrate metabolism, light response, and hormone signaling pathways. This provides key clues for understanding the genetic basis of sweet corn's unique flavor.
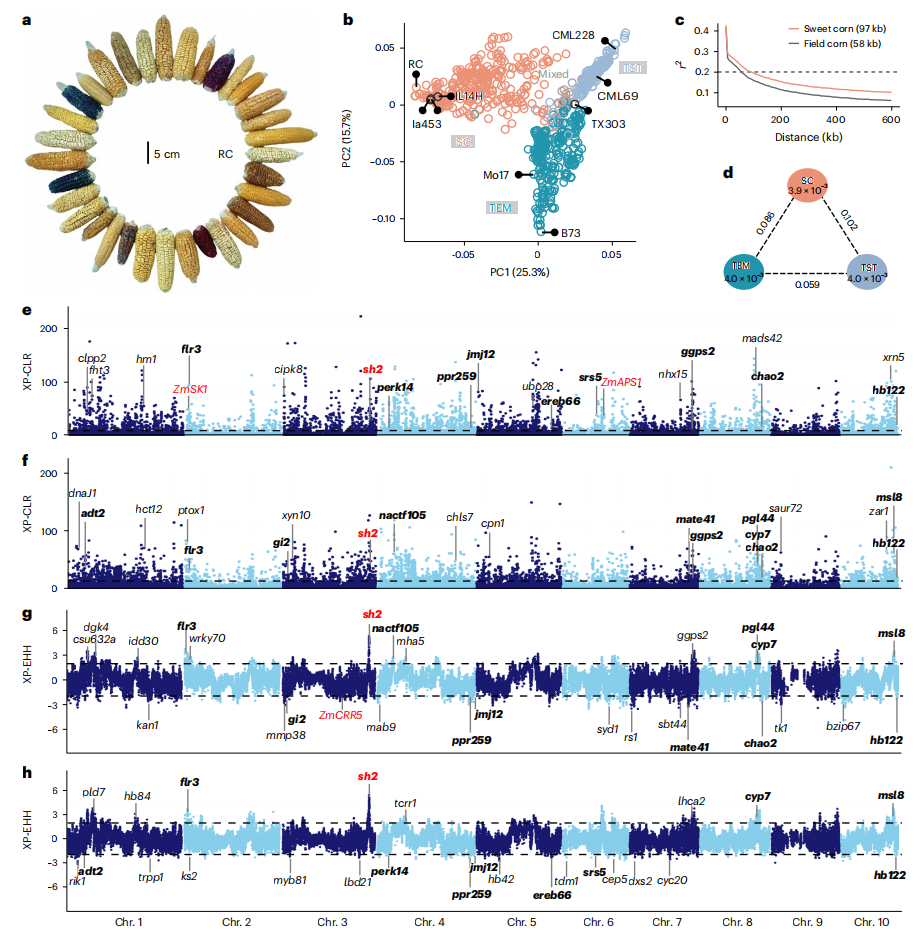
Figure 1. Genetic diversity between sweet-corn and field-corn populations. (Li, et al. 2025)
Further analysis revealed that sweet corn flavor is a complex trait comprised of multiple dimensions, including sweetness, pericarp thickness, crispness, and aroma. Surprisingly, improving flavor scores does not necessarily require increasing sugar content—reducing pericarp thickness and improving texture can also significantly enhance the flavor experience. This discovery challenges traditional views and provides new perspectives and directions for subsequent quality improvement. Based on the aforementioned multi-omics big data, the research team identified three novel flavor-regulating genes: ZmAPS1 regulates adenosine metabolism, affecting the synthesis of flavor precursors; ZmSK1 affects quinic acid synthesis and is closely related to sweet corn aroma; ZmCRR5 regulates fructose accumulation, playing a crucial balancing role between sweetness and yield. Further research confirmed that these genes play regulatory roles in different metabolic pathways, acting as three precise "conductors" in the flavor formation process, jointly regulating the edible quality of sweet corn.
This study innovatively integrates genomic (DNA information), transcriptomic (gene expression information), and metabolomic (small molecule compound information) data to construct a precise predictive model for sweet corn flavor. Analysis shows that, based on genome-wide single nucleotide polymorphism (SNP) data, 42.0% to 73.7% of the differences in flavor scores can be explained. Researchers further identified 4711 genes significantly associated with flavor and 139 key flavor metabolites. Among them, sucrose, norborneol, and heptanoic acid are positively correlated with flavor; while maltose, xylose, and allantoin are negatively correlated with flavor. These findings provide valuable genetic resources and metabolite markers for flavor improvement, contributing to the shift in maize breeding from empirical selection to precision design.
Another core breakthrough of this study lies in revealing the intrinsic trade-off mechanism between flavor and yield, and proposing a new breeding approach of "flavor design." Through multi-omics analysis, the team identified a group of key genes and metabolites that can simultaneously regulate yield and flavor formation. For example, in natural populations, the ZmCRR5 gene exhibits multiple variant types (alleles), with one relatively low-frequency (approximately 13%) allele possessing both high yield and excellent flavor. The discovery of such favorable alleles provides a direct target for breeding new varieties.
This research provides an important example of the transformation of crop breeding from a "yield-oriented" approach to one that emphasizes both quality and yield. It is the first to systematically reveal the genetic interaction mechanism between sweet corn flavor and yield, particularly identifying candidate genes that can enhance flavor while maintaining or even increasing yield, providing a scientific basis for achieving the goal of "synergistic improvement of flavor and yield." Furthermore, the constructed flavor prediction model can predict flavor performance based on genomic information, laying a technical foundation for molecular breeding (such as gene editing and marker-assisted selection), and is expected to significantly improve breeding efficiency and accuracy.
In the future, the research team plans to combine gene editing with marker-assisted selection technology to accelerate the breeding of high-yielding and high-quality new sweet corn varieties. This research not only provides technological support for the upgrading of the sweet corn industry, but its multi-omics integrated research framework and technical approach also provide important reference for flavor improvement in other fruit and vegetable crops.