Comprehensive Plant Protein Profiling Solutions
Understanding the dynamic protein landscape of plants is crucial for uncovering the molecular mechanisms governing growth, stress responses, and metabolic regulation. Our proteomics analysis services provide end-to-end solutions for identifying, quantifying, and characterizing plant proteins at a systems level.
From sample preparation to data interpretation, we combine cutting-edge mass spectrometry (MS) platforms with bioinformatics expertise to deliver reproducible, high-resolution proteomic profiles. Whether you need to monitor differential protein expression between treatments, dissect complex signaling networks, or map post-translational modifications, our customized workflows help you transform raw data into actionable biological insights.
Lifeasible's quantitative and comparative proteomics services include both labeled and label-free proteomics approaches, each customizable to meet diverse research needs and challenges.
Quantitative and comparative proteomics are core technologies for analyzing protein composition, dynamics, and functional interactions across diverse biological environments. This approach enables researchers to study protein expression patterns, identify differentially expressed proteins, and understand the complex mechanisms underlying biological processes. Through both labeled and label-free approaches, we provide high-quality, cost-effective, and reliable services to promote advanced agricultural research and innovation.
![]()
Plant Tissue Collection and Protein Extraction
![]()
Protein Digestion & Optional Isotope/Chemical Labeling
![]()
LC-MS/MS Analysis
![]()
Data Processing & Quantification
![]()
Bioinformatics & Functional Insights
Our labeled proteomics services utilize advanced stable isotope and chemical labeling technologies for in vivo and in vitro analysis. This approach ensures precise protein quantification, enhancing the sensitivity and reproducibility required for analyzing complex samples.
Applications
Our label-free proteomics service provides a technically simple yet powerful protein analysis method using mass spectrometry for direct quantification without labeling. This method preserves proteins in their native state, providing reliable data for characterizing proteins and screening for differentially expressed proteins under specific conditions.
Applications

Lifeasible provides cutting-edge targeted proteomics analysis services, leveraging advanced technologies to quantitatively and sensitively measure specific target proteins in complex biological samples. This service is crucial for agricultural research, providing precise protein data to understand crop growth, development, and stress responses.
Targeted proteomics utilizes parallel reaction monitoring (PRM), a technique that relies on high-resolution, high-accuracy mass spectrometry. This approach selectively detects and accurately quantifies target proteins, overcomes background interference, and reduces false positives. PRM eliminates the need for pre-set transitions, enabling comprehensive product ion scanning and analysis across a wide linear range.
PTMs such as phosphorylation, glycosylation, and ubiquitination are key regulators of protein function, stability, and interaction networks. Lifeasible's PTM analysis service utilizes advanced enrichment techniques and high-resolution mass spectrometry to comprehensively and sensitively detect modifications across a wide range of plant proteins. This enables researchers to elucidate complex regulatory mechanisms during plant signaling and development.
Protein-protein interactions are central to the assembly of functional complexes and signaling pathways. Lifeasible offers comprehensive PPI mapping services to identify and characterize the interacting partners of plant proteins, revealing their functional networks and regulatory roles. By combining classical and advanced proteomics approaches, we provide reliable and reproducible data for both stable and transient interactions.
Understanding the spatial distribution of proteins within plant cells is crucial for interpreting their functions. Lifeasible offers specialized subcellular and organellar proteomics services to analyze proteins within chloroplasts, mitochondria, plasma membranes, nuclei, and other organelles. These services provide detailed insights into the compartmentalization of plant metabolic and regulatory processes.

High-quality proteomics data requires specialized computational analysis to extract meaningful biological insights. Lifeasible's bioinformatics and data interpretation services provide comprehensive analysis of plant proteomics datasets, ensuring that complex data is transformed into clear, actionable conclusions.
![]()
Experimental Design and Consultation
We collaborate with you to refine hypotheses, select appropriate proteomics strategies, and define experimental replicates.
![]()
Sample Preparation
Plant tissue is homogenized, proteins are extracted and quantified, and then digested to generate peptides.
![]()
Fractionation & Enrichment (Optional)
Peptides can be fractionated or enriched to identify specific PTMs for increased analytical depth.
![]()
Mass Spectrometry Acquisition
Peptide mixtures are analyzed using high-resolution LC-MS/MS instrumentation with optimized settings for the plant proteome.
![]()
Data Processing & Bioinformatics
Advanced software identifies and quantifies proteins, performs statistical analysis, and annotates biological functions.
![]()
Comprehensive Reporting
Deliverables include detailed protein lists, differential expression analysis, and pathway/network insights.
Our streamlined workflow integrates best practices at every stage to ensure data quality and reproducibility.
To obtain reliable and reproducible results, please follow these guidelines.
| Requirement | Guidelines |
| Sample Type | Fresh or flash-frozen plant tissue (leaves, roots, stems, seeds, fruit). Avoid using desiccated or degraded material. |
| Quantity | For most plant species, at least 0.5-1 g of tissue (wet weight) is required per biological replicate. |
| Replicates | At least three biological replicates per experimental condition are strongly recommended. |
| Pre-treatment | Remove excess soil or contaminants; avoid the use of chemical preservatives. |
| Storage & Transport | Snap-freeze in liquid nitrogen immediately after harvest; store at -80°C and transport on dry ice. Avoid repeated freeze-thaw cycles. |
Plant-Focused Expertise
Extensive experience with diverse plant species, from model organisms like Arabidopsis to genomically complex crop species.
Flexible Quantitation Strategies
Choose between labeled or label-free workflows based on your research objectives and sample limitations.
High Sensitivity & Accuracy
Advanced LC-MS/MS systems ensure robust detection of low-abundance proteins.
End-to-End Support
From experimental design to biological interpretation, we provide expert guidance and full technical support.
Ready to accelerate your plant proteomics analysis?
Contact us to request a free consultation, get a custom quote, or discuss your experimental goals directly with one of our technical experts.
Proteomics is the large-scale study of all proteins in a cell, tissue, or organism under specific conditions. In plants, proteomics provides a direct link between genotype and phenotype, helping to understand how proteins respond to environmental factors, developmental stages, and stress conditions. By combining advanced mass spectrometry techniques with bioinformatics, proteomics can simultaneously identify and quantify thousands of proteins.
Transcriptomics reveals gene expression patterns, while proteomics captures functional outputs—the proteins that carry out cellular processes. In plant science, proteomics helps uncover the molecular mechanisms behind crop yield, stress tolerance, disease resistance, and nutrient utilization. These insights are crucial for precision breeding, crop improvement, and sustainable agriculture.
Typically 100–200 µg of total protein per biological replicate is recommended, though requirements may vary depending on the complexity of the analysis.
Yes. We employ optimized extraction protocols and cleanup steps to remove interfering compounds.
Standard projects are usually completed within 6–8 weeks, depending on the complexity of the experimental design.
Yes. Our team delivers comprehensive data interpretation, including differential expression analysis, pathway mapping, and visualization.
Labeled proteomics is well-suited for experiments requiring high-precision multiplexing and direct comparisons across multiple samples. Label-free methods are more flexible and cost-effective for large-scale or exploratory projects. Our experts can help you select the most appropriate method based on your objectives, sample type, and budget.
Yes. Our plant proteomics platform supports the detection of a wide range of PTMs, including phosphorylation, glycosylation, ubiquitination, and acetylation.
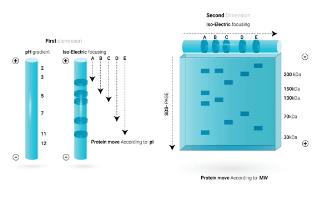
Separation of Proteins by Two-dimensional Gel Electrophoresis

Determining The Relative Molecular Mass of Proteins Using SDS-PAGE

Separation of Serum Proteins by Cellulose Acetate Membrane Electrophoresis

Extraction of Total Protein from Wheat Leaves