Transcriptomics is a powerful approach for understanding gene expression and regulation at the RNA level. It encompasses the identification and quantification of all RNA molecules transcribed from a genome under specific biological or environmental conditions.
In plants, transcriptomics studies have become integral to elucidating the molecular mechanisms underlying growth, development, stress responses, and metabolite biosynthesis. By comparing transcriptional profiles across treatments or developmental stages, researchers can identify candidate genes associated with desirable traits such as yield increase, drought tolerance, or disease resistance.
Lifeasible's plant transcriptomics analysis service combines advanced sequencing technologies with rigorous bioinformatics pipelines to provide comprehensive, accurate, and biologically meaningful results. Whether you are studying changes in gene expression or transcriptional regulation in crops, our team offers end-to-end solutions to accelerate your plant transcriptomics research.
RNA sequencing (RNA-Seq) provides a comprehensive snapshot of the transcriptome, enabling accurate quantification of gene expression levels and discovery of novel transcripts. This method is widely used to study gene expression dynamics, identify differentially expressed genes, and investigate molecular mechanisms in plants under various physiological or environmental conditions.
![]()
RNA Extraction & Quality Control
Isolate high-integrity total RNA and assess its concentration, purity, and integrity.
![]()
Library Preparation
Enrich mRNA or deplete rRNA, followed by cDNA synthesis and adapter ligation.
![]()
Sequencing
Libraries are sequenced to generate paired-end reads.
![]()
Data Processing
Reads are filtered, mapped to the reference genome, and quantified for gene expression.
![]()
Differential Expression and Functional Analysis
Identification of significant DEGs and pathway-level interpretation.
De novo transcriptome assembly technology enables transcriptome analysis of plant species lacking a reference genome. It reconstructs full-length transcripts from short-read data, providing insights into gene structure, expression, and functional diversity.
![]()
RNA Isolation and Quality Control
Extraction of total RNA from plant tissues with strict quality control.
![]()
cDNA Library Construction
Preparation of libraries optimized for transcript diversity and coverage.
![]()
High-throughput Sequencing
Generation of sufficient read depth for accurate transcript reconstruction.
![]()
De Novo Assembly
Short reads are assembled into contigs and unigenes using established algorithms.
![]()
Functional Annotation
Transcripts are annotated against multiple databases to assign biological functions.
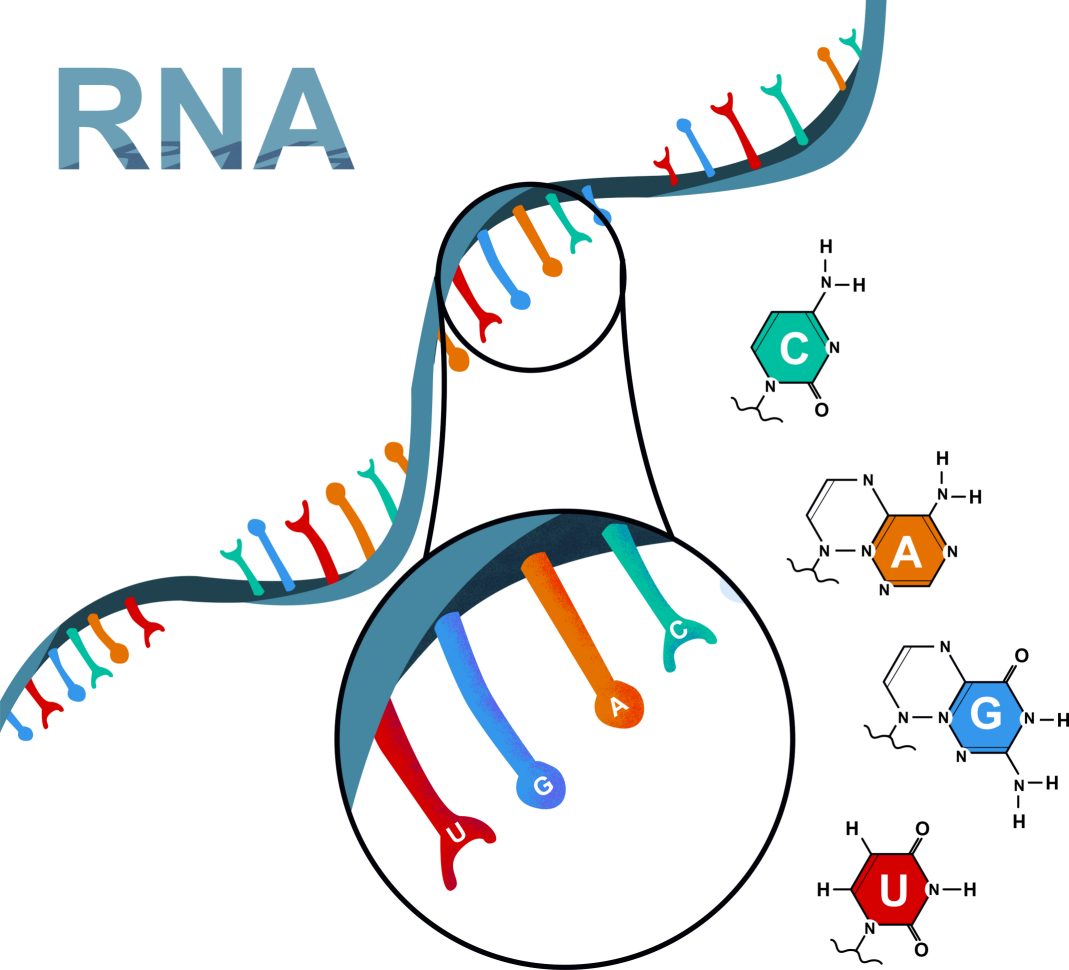
Full-length RNA sequencing (Iso-Seq) captures the complete transcript isoforms without assembly, enabling direct visualization of alternative splicing events, gene fusion transcripts, and long non-coding RNAs. This approach provides a comprehensive understanding of transcript diversity and structural complexity.
Small RNA sequencing enables comprehensive profiling of microRNAs (miRNAs), small interfering RNAs (siRNAs), and other noncoding RNAs that regulate plant gene expression. This is crucial for exploring post-transcriptional regulation, stress adaptation, and developmental control.

We provide comprehensive bioinformatics analysis, from raw sequencing to biological insights, to interpret transcriptome data. This analysis integrates differential expression, functional enrichment, and network modeling to reveal transcriptional regulation and pathway involvement.
![]()
Sample Collection and RNA Extraction
Fresh or frozen plant tissue samples (e.g., leaves, roots, flowers) are processed using optimized protocols to obtain high-quality RNA.
![]()
Library Preparation
Poly(A) enrichment or rRNA depletion, followed by cDNA synthesis and adapter ligation.
![]()
Sequencing
Sequencing is performed based on research objectives.
![]()
Data Preprocessing
Reads undergo quality control, trimming, and alignment or de novo assembly.
![]()
Bioinformatics Analysis
Differential expression, functional enrichment, and co-expression network analysis are performed.
![]()
Report Generation
Clients receive a detailed analysis report containing raw data, visualization charts, and biological interpretation.
Our streamlined workflow ensures efficient, high-quality plant transcriptomics results, from RNA extraction to report generation.
| Requirement | Guidelines |
| Sample Type | Fresh or frozen plant tissue (leaves, roots, stems, seeds, flowers, callus, or suspended cells). |
| RNA Sample |
|
| Tissue Sample | ≥200 mg per sample if RNA extraction is performed by our lab. |
| Storage and Shipping | Remove excess soil or debris; avoid the use of chemical preservatives. |
| Storage & Transport |
|
Customizable Workflows
Customizable workflows for model and non-model plant species.
Experienced Bioinformatics Support
Provides detailed data interpretation tailored to experimental objectives.

High Sensitivity & Reproducibility
Ensures accurate quantification of low-abundance transcripts.

Complete Service Package
From sequencing to in-depth analysis to publication-ready results.
Ready to advance your plant transcriptomics research?
Contact Lifeasible now for a free consultation and a custom-designed sequencing quote.
| Transcriptomics Service Focus | Core Service Capabilities | Key Applications in Plant Research |
| Eukaryotic Transcriptome Sequencing |
|
|
| Prokaryotic Transcriptome Sequencing |
|
|
| Transcriptome Analysis of Plant Protoplast Reprogramming into Stem Cells |
|
|
Transcriptomics is the systematic study of the transcriptome (the complete set of RNA transcripts produced by an organism's genome during a specific developmental stage or environmental conditions). It serves as a crucial intermediary, directly measuring which genes are active and how much they are expressed, connecting the static blueprint of the genome with the dynamic realization of the proteome. It goes beyond simple gene identification to provide a real-time understanding of their regulation and function.
The field has undergone a dramatic transformation with the adoption of next-generation sequencing (NGS) technologies, primarily RNA-Seq. Unlike traditional methods such as microarrays, RNA-Seq offers:
Transcriptomics is crucial for unraveling the complex mechanisms unique to plant life. It enables researchers to:
Transcriptomics data is most powerful when it serves as the foundation for systems biology approaches. As the causal "input" in multi-omics studies, it enables researchers to:
Yes. We accept both extracted RNA and frozen tissue. If RNA extraction is required, we use plant-specific protocols to ensure purity and integrity.
Our team will assess sample quality and provide feedback before library construction. We can perform re-extraction or optimization if necessary.
Typically, 20 to 40 million reads per sample is sufficient to cover most differential expression studies. Deeper sequencing depth is recommended for isoform or de novo assembly.
The standard turnaround time is 4-6 weeks, depending on the project size and analysis complexity.
Yes, our bioinformatics team provides comprehensive interpretation and visualization services to help you understand and publish your research results.

Detection of Arabidopsis SNP Genetic Markers

SSR Identification of Cotton Salt Tolerance

RAPD Analysis of Genetic Diversity for Salt Tolerance in Wheat

AFLP Analysis of Genetic Diversity and Genetic Relationships of Spinach Germplasm