On April 22, 2025, Steven E. Jacobsen's team at the University of California at Los Angeles published a research paper titled "Viral delivery of an RNA-guided genome editor for transgene-free germline editing in Arabidopsis" in Nature Plants. This paper demonstrates complete viral delivery of CRISPR/Cas components in plants, achieving heritable genome editing in Arabidopsis. Notably, one of the authors of the paper is Jennifer A. Doudna, a Nobel laureate in CRISPR/Cas.
Genome editing technology is revolutionizing plant biology, enabling diverse applications through precise DNA modification. However, delivering editing systems into plant cells remains a challenge. Traditional methods, such as tissue culture and transgenic techniques, are often time-consuming, limited to specific plant species and genotypes, and can also lead to unintended genomic and epigenomic changes. Although plant viruses naturally infect and spread to most plant tissues, making them potential delivery systems for editing agents, many viruses have limited ability to express exogenous proteins, making them difficult to carry large CRISPR-Cas systems. Therefore, developing a novel genome editing platform that overcomes traditional delivery barriers is crucial for accelerating plant biotechnology and basic research.
The research team selected the ultracompact RNA-guided endonuclease TnpB (e.g., ISYmu1) and its guide RNA. These enzymes have a small molecular weight (approximately 400 amino acids), making them suitable for expression in plant viral vectors.
Tobacco rattle virus (TRV) was modified to carry the TnpB enzyme and guide RNA. Two TRV2 architectures (Architecture_A and Architecture_B) were constructed, and a tRNAIleu sequence was incorporated to facilitate systemic propagation.
Both architectures were delivered via Agrobacterium infiltration, eliminating the need for tissue culture.
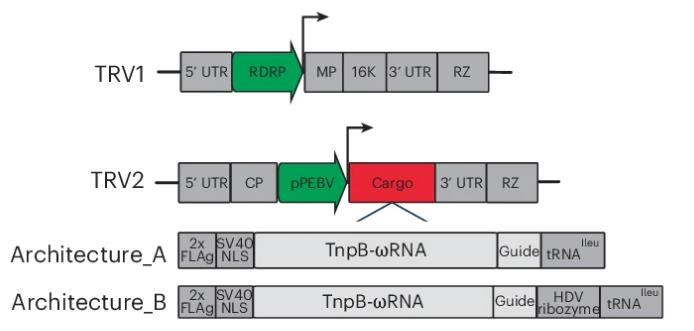
Figure 1. Schematic of the TRV1 and TRV2 plasmids. (Weiss, et al. 2025)
The editing efficiency of TnpB enzymes, including ISYmu1, was tested in Arabidopsis protoplasts. Results showed that ISYmu1 had high editing efficiency.
Arabidopsis plants expressing ISYmu1 were generated using transgenic technology, and their editing efficiency was evaluated under different temperatures and genetic backgrounds.
ISYmu1-mediated genome editing was achieved in Arabidopsis using a modified TRV viral vector, and the editing efficiency and specificity were evaluated.
For the AtPDS3 gene (mutations leading to an albinism phenotype), Architecture_B combined with heat shock treatment achieved an editing efficiency of 3.3% in wild-type Arabidopsis and as high as 8.9% in the ku70 mutant (optimized DNA repair).
For the AtCHLI1 gene (mutations leading to etiolation), heat shock treatment increased the editing efficiency to 18.4%, resulting in distinct etiolation.
Among the offspring of AtPDS3-edited plants, 18%-43% showed bi-allelic editing (albino seedlings), and 8.5% showed mono-allelic editing. RT-PCR confirmed the absence of TRV virus residues in the offspring, achieving transgene-free editing.
Whole-genome sequencing analyzed the off-target effects of ISYmu1-mediated editing, demonstrating high target site specificity.
Direct viral delivery bypasses tedious transformation steps and is applicable to a wide range of plants.
TRV infects over 400 plant species (such as tomato and potato), potentially covering a wide range of crops.
Chimeric tissues can be created to study the function of embryo-lethal genes.
This study successfully combined the ultra-compact TnpB enzyme with a tobacco rattle virus vector to achieve transgene-free genome editing in Arabidopsis. This approach overcomes the limitations of traditional delivery systems, providing a rapid, efficient, and easy-to-use platform for plant genome editing. Furthermore, the study confirmed the high editing efficiency and specificity of the ISYmu1 enzyme in plants, as well as the heritability of the editing.
Utilizing ultra-compact RNA-guided endonucleases such as TnpB overcomes the limited capacity of plant viral vectors.
By modifying the TRV viral vector, efficient delivery of the TnpB enzyme and its guide RNA was achieved, providing a new delivery system for plant genome editing.
The ISYmu1 enzyme demonstrated high editing efficiency and specificity in Arabidopsis, and the editing results were stably inherited in subsequent generations.
This method is not only applicable to Arabidopsis but also has the potential to be expanded to other plant species, providing a powerful tool for plant biotechnology and basic research.