Proteomics Analysis of Plant Fungal
When infested by foreign fungi, plants will achieve sensing, transmission, and biological defense against fungal infestation signals by changing the expression pattern of their proteome or the activity of enzymes and other lifestyles. Proteins have specific activity patterns, such as protein structure formation, modification and processing, translocation and localization, and protein-protein interactions, which usually cannot be reflected directly from genomic information. The rapid development and refinement of proteomics technologies provide a range of powerful tools to directly study gene function at the protein level in a high-throughput manner. We can help our customers explore the pathogenesis and metabolic network structure of plant fungi using conventional proteomics research methods.
Lifeasible can use proteomic methods to study the process of fungal infestation of plants, analyze the relevant genes and proteins, and help customers explore the mechanisms of plant-fungal interactions at the molecular level.
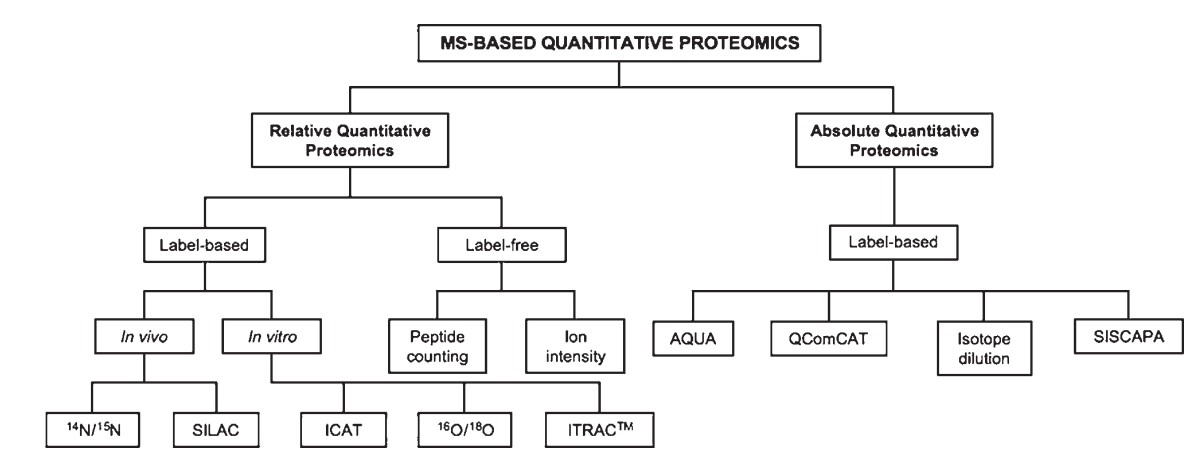 Figure 1. Scheme of quantitative proteomic methods. (Gonzalez-Fernandez R, et al., 2012)
Figure 1. Scheme of quantitative proteomic methods. (Gonzalez-Fernandez R, et al., 2012)
- Differential proteomics analysis of plant-fungal interactions. We can compare the differences in proteomic expression profiles of fungal-infested plant tissues and corresponding normal tissues, analyze and identify intentional functional proteins associated with fungal induction, understand the differential expression of different functional proteins and their functions under various biotic stresses, and thus elucidate the molecular mechanisms of fungal pathogenesis or host plant disease resistance.
- Differentially expressed functional proteins analysis of plant hosts and pathogenic fungi. Plant hosts can trigger basic resistance in plant hosts by sensing pathogenic infestation signals, such as pathogen-associated molecular patterns associated with pathogenicity, cellular degradation enzymes, and other signaling factors for induced expression. We can use proteomics to analyze the functional proteins of the plant host during plant-pathogen interactions and identify pathogenesis-related and defense response-associated proteins.
- ITRAQ-based proteomics analysis. Isotope labeling relative and absolute quantitative techniques can screen differential protein profiles between different samples. We can dynamically and quantitatively analyze the differential protein expression profiles in different states of fungal physiology and pathology to help our customers analyze the pathogenic mechanism of fungi and explore the interaction between fungi and plant hosts.
The proteomics analysis techniques we can provide are listed below.
| Isotope labeling |
Two-dimensional polyacrylamide gel electrophoresis |
| Mass spectrometry |
Peptide sequence labeling |
| Protein microarrays |
Nuclear magnetic resonance techniques |
Lifeasible works on differential proteome expression during plant-fungal interactions to help customers understand the mechanisms of plant fungal infestation. As your trusted partner, we can meet all your fungal proteomics analysis needs and provide you with efficient and high-quality services. If you want to know the details, please contact us.
Reference
- Gonzalez-Fernandez R, Jorrin-Novo JV. Contribution of proteomics to the study of plant pathogenic fungi. J Proteome Res. 2012 Jan 1; 11(1): 3-16.
For research or industrial raw materials, not for personal medical use!
 Figure 1. Scheme of quantitative proteomic methods. (Gonzalez-Fernandez R, et al., 2012)
Figure 1. Scheme of quantitative proteomic methods. (Gonzalez-Fernandez R, et al., 2012)