Fungal species are huge and diverse, among which there are more than 8,000 species of phytopathogenic fungi that can cause more than 30,000 kinds of plant diseases. The common smut, powdery mildew, rust, downy mildew, etc., on plants are caused by fungi. In recent years, the study of the pathogenic mechanism of plant pathogenic fungi has also become one of the hot issues in the field of plant pathology, and understanding the mechanism of interactions between plants and pathogenic fungi is of great significance for the effective control of pathogenic fungal damage and selection of disease-resistant varieties.
Lifeasible has extensive experience in the isolation and identification of plant pathogenic fungi and can help customers achieve the identification of pathogenic fungi using DNA molecular marker technology.
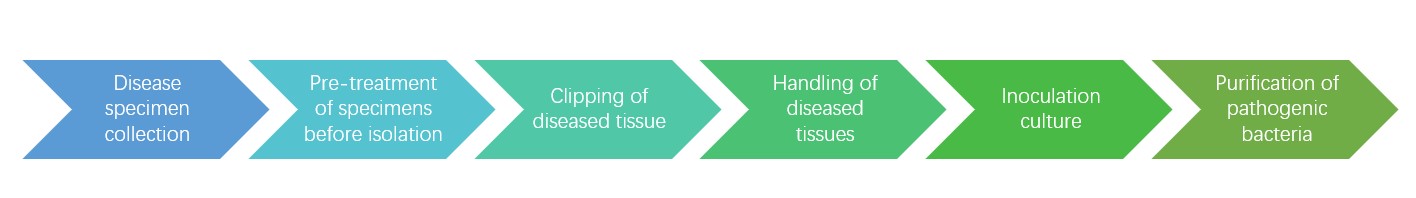 Figure 1. Plant pathogenic fungal isolation process.
Figure 1. Plant pathogenic fungal isolation process.
Lifeasible can help customers isolate and identify plant pathogenic fungi using various molecular techniques based on DNA data. As your trusted partner, we can meet your needs for mycorrhizal fungi identification and isolation services and provide efficient and high-quality services. If you want to know the details, please contact us.
Reference
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024
Application of Exosomes in Facial Beauty
April 12, 2024