Salt stress is threatening the production of many crops globally and is a major abiotic stress that affects plant growth and development and reduces yield and productivity. With the alarming increase in global warming, salinity conditions are rapidly increasing, especially in arid and semi-arid regions. Salinity is often recognized as an excessive accumulation of sodium ions in the soil, leading to osmotic stress and ionic toxicity. These two major effects of salt damage result in reduced photosynthetic efficiency, redistribution of cell wall components, reduced cell expansion and division, and oxidative damage due to reactive oxygen species (ROS). In response to salt stress, plants activate various tolerance mechanisms, including physiological, biochemical, and molecular changes. These different mechanisms are primarily mediated through sensing and signaling pathways, strong transcriptional reprogramming, hormone osmotic agents and ion homeostatic stabilization, increased reactive oxygen species scavenging, and activation of transport and photosynthetic systems.
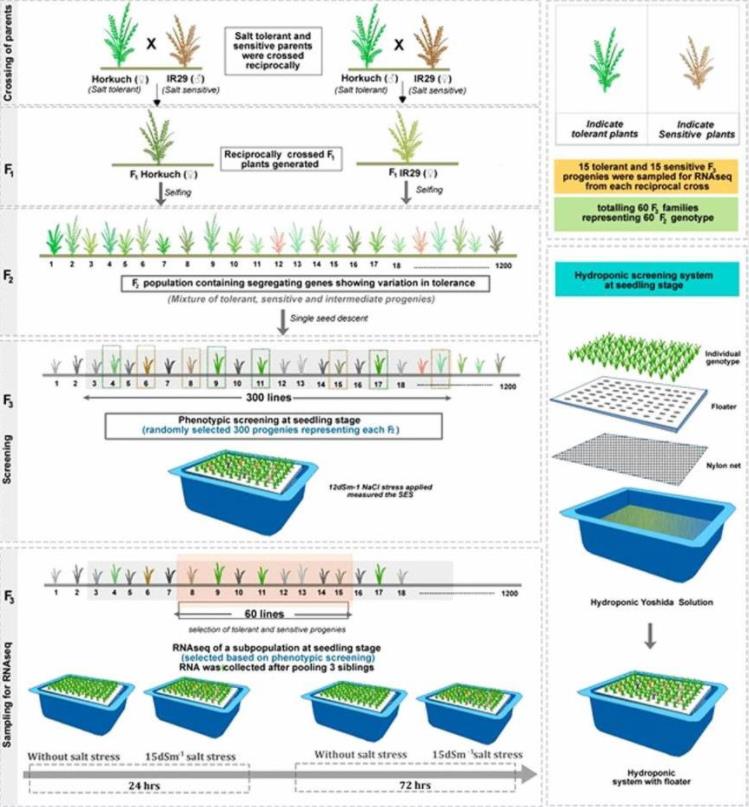 Fig. 1. Gene Expression analysis associated with salt stress in a reciprocally crossed rice population. (Razzaque et al., 2019)
Fig. 1. Gene Expression analysis associated with salt stress in a reciprocally crossed rice population. (Razzaque et al., 2019)
Salinity is one of the most pressing abiotic stressors threatening plant growth and agricultural production worldwide. As one of the leading service providers in plant biotechnology, Lifeasible offers professional solutions to analyze salt stress response genes. Through detailed functional analysis of these genes, we aim to help our clients with gene cloning, transformation, gene editing, and developing salt-resistant crop varieties that maintain productivity and quality.
We offer a variety of methods to analyze salt-stress-responsive genes, including but not limited to:
We offer microarray analysis and marker-based sequencing methods to analyze various plants' overall gene expression profiles and create genetic maps of crop salt-stress-responsive genes. Genetic mapping allows us to provide important gene families that significantly impact salt stress-related plant development. In addition, we provide detailed studies of identified genes, such as phylogenetic analysis, gene structure, subcellular localization, possible gene ontology-based functions, miRNAs, cis-elements, and expression profiles in leaves and roots.
RNA transcript analysis is a powerful technology for genome-wide transcript characterization, differential gene expression analysis, variant detection, and gene-specific expression. We offer RNA-seq to characterize salt stress-induced transcripts in a variety of plants, such as barley (Hordeum vulgare L.), sweet potato (Ipomoea batatas (L.) Lam.), canola (Brassica napus L.), and Arabidopsis. In addition, we can use this technique to analyze leaf and root differentially expressed genes under salt stress and to identify differentially expressed genes (DEGs) encoding several transcription factors and signal transduction components associated with salt tolerance. These candidate genes include ABA response elements, calcium signaling, LRR-RLK, protein kinases, several transcription factors (MYB, bHLH, HD-ZIP, WRKY, MADS-box, NAC families), antioxidant genes, etc.
Lifeasible provides complete information on key genes associated with salt tolerance, essential for future studies of the specific molecular mechanisms underlying plant salt stress responses. The identified candidate genes provide a valuable resource for future genetic engineering studies of crops to develop new varieties with more salt-tolerant traits and will be used to establish effective applied breeding programs. Contact us today to learn more about our solutions.
Reference