With the continuous development of gene sequencing technology, more and more fungi have completed whole genome code-breaking. In the fungal evolution process, each species has developed unique genetic traits due to natural and anthropogenic selection and other influences. Therefore, a single genome is far from covering all genetic sequences of the entire species. Fungal pan-genome refers to the core genome that exists in almost all individuals of a fungal species and the variable genome that exists in only some individuals. Lifeasible can use fungal pan-genome analysis to obtain more accurate and comprehensive variation information, analyze fungal strain evolution, adaptation, and community structure, and screen for functional genes or functional sequences in variable genomes.
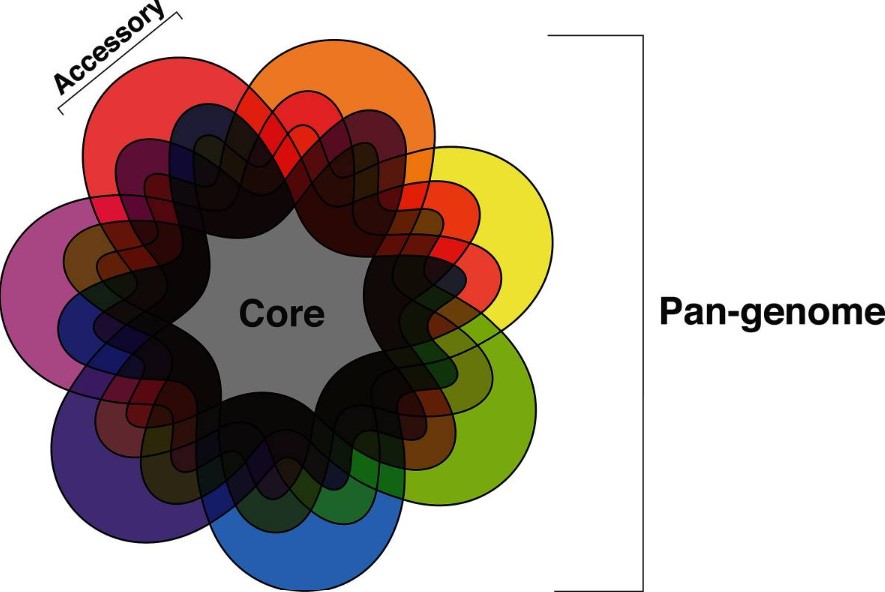 Figure 1. Venn diagram of the pan-genome. (McCarthy CGP et al., 2019)
Figure 1. Venn diagram of the pan-genome. (McCarthy CGP et al., 2019)
Lifeasible can provide fungal pan-genome analysis to help customers investigate the important role of variable genes in the pathogenicity of plant pathogenic fungi and address questions related to plant-pathogenic fungal coevolution through pan-genome studies. As your trusted partner, we can meet your fungal phylogenetic analysis needs and provide you with efficient, high-quality services. If you want to know the details, please contact us.
Reference
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024
Application of Exosomes in Facial Beauty
April 12, 2024