Promoter substitution is using a new promoter to replace the original promoter of a silenced gene, thus allowing normal expression of the silenced gene in the original host. Since the secondary metabolites of fungi may be toxic to the original host, researchers often use inducible promoters to replace the original promoters of silenced genes to not affect the fungus's growth and activate the silenced genes to obtain new active natural products. However, when performing promoter substitution, the promoters of all silenced biosynthetic genes in the gene cluster need to be replaced, and the workload is high. Lifeasible is committed to developing promoter substitution-mediated activation strategies for silent gene clusters to help customers achieve efficient utilization of fungal resources.
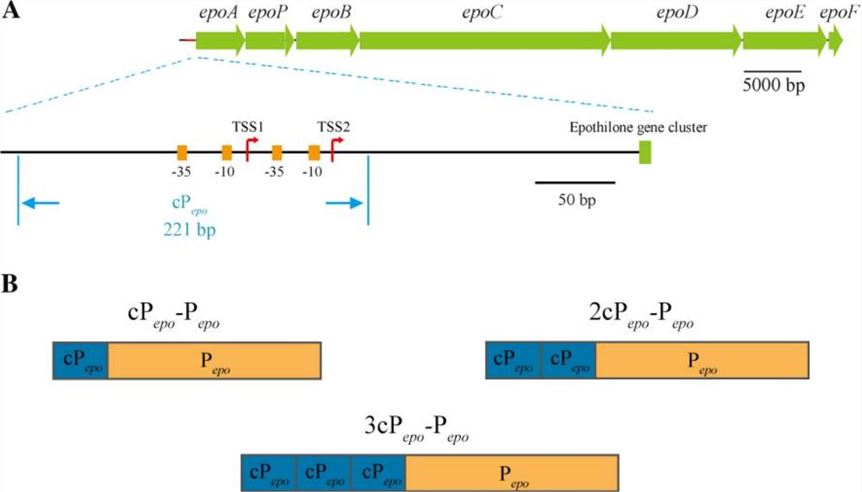 Figure 1. Physical map of the epothilone biosynthetic gene cluster and the promoter Pepo. (Van Dijck P, et al., 2017)
Figure 1. Physical map of the epothilone biosynthetic gene cluster and the promoter Pepo. (Van Dijck P, et al., 2017)
The specific services we can provide are as follows. Including but not limited to.
| Promoter cloning |
|
| Promoter activity analysis |
|
| Analysis of promoter core elements |
|
| Genetic editing |
|
| Dual fluorescence reporting system |
|
The commonly used eukaryotic promoters are as follows.
| Eukaryotic promoters | Types of transcribed RNA | Expression features | Other features |
| CMV | mRNA | Constitutive expression | May contain an enhancer that will silence in some cell types |
| EF1a | mRNA | Constitutive expression | Stable expression in all cell types and physiological states |
| SV40 | mRNA | Constitutive expression | May contain an enhancer |
| CAG | mRNA | Constitutive expression | Contains CMV enhancer |
| UAS | mRNA | Specific expression | Requires the presence of the Gal4 gene to activate the promoter |
| Ac5 | mRNA | Constitutive expression | Expression systems commonly used in Drosophila |
| TEF1 | mRNA | Constitutive expression | Similar to the mammalian EF1α promoter |
| GDS | mRNA | Constitutive expression | Great ability to start expressions |
| ADH1 | mRNA | Inhibition with ethanol | This promoter has a strong initiation ability and high expression level when it is full-length; it is constitutively expressed when it is truncated and has a low expression level |
| … | … | … | … |
Lifeasible can provide full operational services for promoter substitution-mediated activation of silent gene clusters, helping our customers to reduce project time. As your trusted partner, we can meet all your fungal phylogenetic analysis needs and provide efficient, high-quality services. If you want to know the details, please contact us.
Reference