Genomics and bioinformatics analysis of genome sequences have revealed silent gene clusters that are not expressed under specific growth conditions or are undetectable at low expression levels. These silent gene clusters are called silent natural product biosynthetic gene clusters. Currently, there are two main strategies to activate these silent gene clusters. One is stochastic activation, mainly by optimizing growth conditions, adding chemical mutagens, trace metal ions, provision of exogenous small molecules, ribosome engineering, co-culture with other organisms, etc. The second is targeted activation based on genomic sequences, mainly targeting regulatory genes. Global regulators are a class of regulators that can simultaneously regulate the expression of multiple genes under specific induction conditions. In addition to epigenetic modifying enzymes, global regulators can also regulate multiple genes at the same time. Lifeasible can help customers simultaneously induce multiple gene expressions through global regulatory factors under specific environmental conditions.
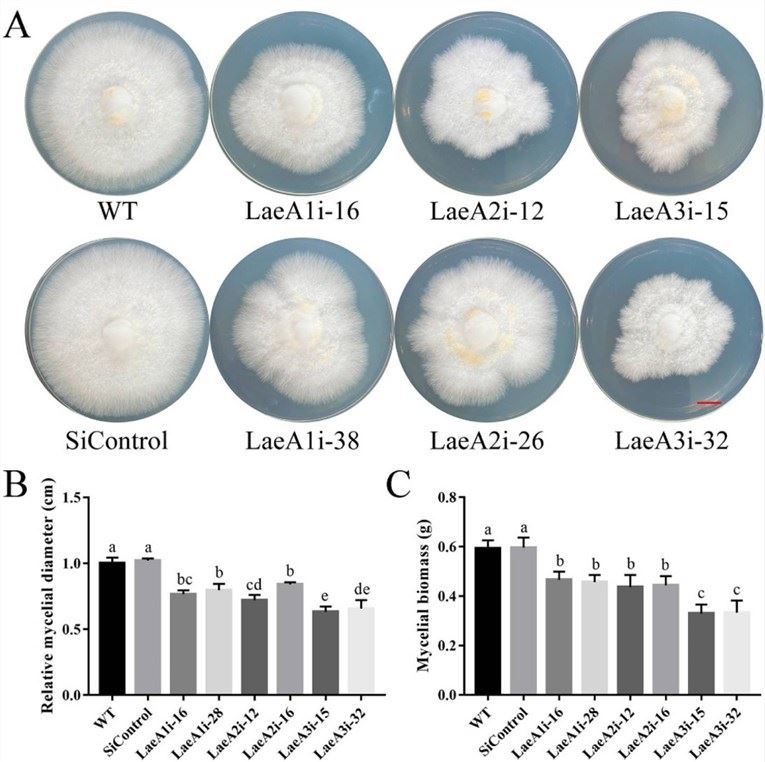 Figure 1. Hyphal growth and biomass of PoLaeA-silenced strains. (Zhang G et al., 2017)
Figure 1. Hyphal growth and biomass of PoLaeA-silenced strains. (Zhang G et al., 2017)
The global regulators currently used for fungal silencing gene cluster activation are listed below, including but not limited to.
Lifeasible can help customers target fungal modification based on gene identification and structural analysis of global transcription factors to activate silent gene clusters and regulate transcriptional expression of secondary metabolite-related genes. As your trusted partner, we can meet all your fungal phylogenetic analysis needs and provide you with efficient and high-quality services. If you want to know the details, please contact us.
Reference
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024
Application of Exosomes in Facial Beauty
April 12, 2024