The need for sustainable energy reserves and enhanced environmental controls have been increasing over the past few years, and this trend appearsto continue. One new weapon to meet this challenge is the development of microalgae. Among these, microalgal proteins contain all essential amino acids and have great potential to become an alternative protein source, with protein concentrates that can be used in the food, feed, and bulk chemical industries. Bioinformatics analysis methods can help elucidate microalgae's molecular mechanisms and thus facilitate their full exploitation. In particular, virtual proteomics represents a rapidly evolving scientific field at the border of bioinformatics and molecular biology, including tools for the prediction, simulation, and modeling of protein complex interactions, as well as insights into transient intracellular signaling pathways and protein evolution.
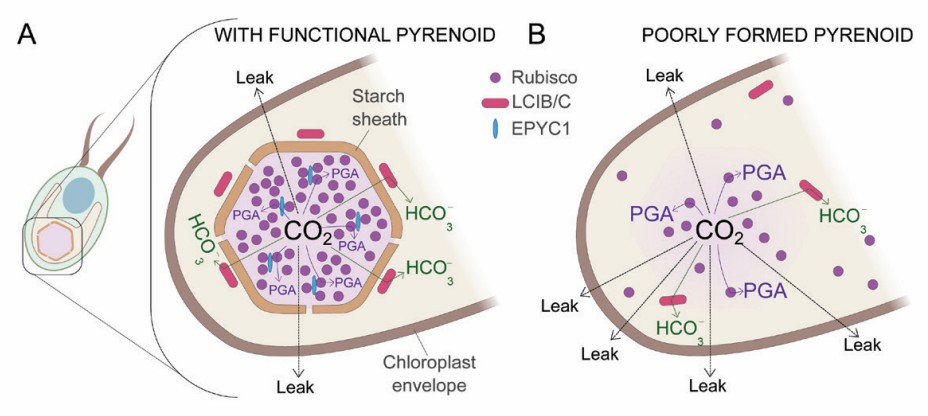 Fig 1. How the pyrenoid reduces CO2 leakage in Chlamydomonas reinhardtii. (Mukherjee A, et al., 2019)
Fig 1. How the pyrenoid reduces CO2 leakage in Chlamydomonas reinhardtii. (Mukherjee A, et al., 2019)
Bioinformatics approaches can now identify putative protein interactions exclusively from genomic sequences. Over the years, our skilled scientists have developed gene fusion and fission-based bioinformatics methods for putative microalgal protein-protein interactions. At Lifeasible, we offer specialized services for the analysis of microalgal protein interactions to predict the 3D structure of microalgal fusion and component proteins and to identify potential protein interactions and functional links in different microalgal species.
The services we can provide include the following:
Our virtual proteomics service applies new bioinformatics methods to the proteomes of five different microalgal organisms, including green algae, Chlamydomonas reinhardtii, Chlorella variegata, Chlorella luminescens, red algae, and diatoms.
Based on bioinformatics analysis of gene fusions and fissions, we can quickly help you predict protein interactions of different microalgae to expand your understanding of microalgal species beyond the analysis of experimental observations. If you are interested in our solutions for the analysis of microalgal protein interactions, please contact us directly.
Reference