Organisms sense diverse environmental signals using complex gene regulatory networks (GRNs) and respond accordingly by coordinating the expression of multiple genes. Cell-free systems (CFS) are an ideal platform for constructing synthetic biological systems because they can rapidly prototype genetic components and pathways. To construct CFSs capable of sensing and responding to diverse environmental signals, new approaches are needed to enhance the capacity of synthetic gene regulatory networks. GRN engineering will promote the development of CFS in the fields of multiple biosensors, biosynthesis platforms, and cell synthesis.
Recently, Diego Alba Burbano from the Center for Synthetic Biology at the University of Washington published a research paper entitled "Engineering activatable promoters for scalable and multi-input CRISPRa/i circuits" on PNAS. This research develops a method for creating high-performing activatable promoters. These promoters can be constructed into a multilayer cascades, parallel cascade, and multi-input CRISPR-activation and interference (CRISPRa/i) GRNs, which provides a general method for studying high dynamic range activatable promoters and realizes a variety of gene regulatory functions in cell-free systems.
First, the researchers characterized the sequences that affect the recruitment of RNA polymerase (RNAP) to synthetically activatable promoters, and their impact on basal and post-activation expression levels. RNAP recruitment is influenced by the affinity of the RNAP sigma subunit (σ) for minimal promoters (-10 and -35 hexamers). In addition, recruitment was also affected by the GC content of the intervening sequence between the −10 and −35 sites. Promoter recognition is enhanced by AT-rich UP elements located upstream of the minimal promoter, which anchor the α-subunit of RNAP. Overall, the sequence composition of these regions affects the recruitment, binding and initiation of transcription of RNAPs at promoters.
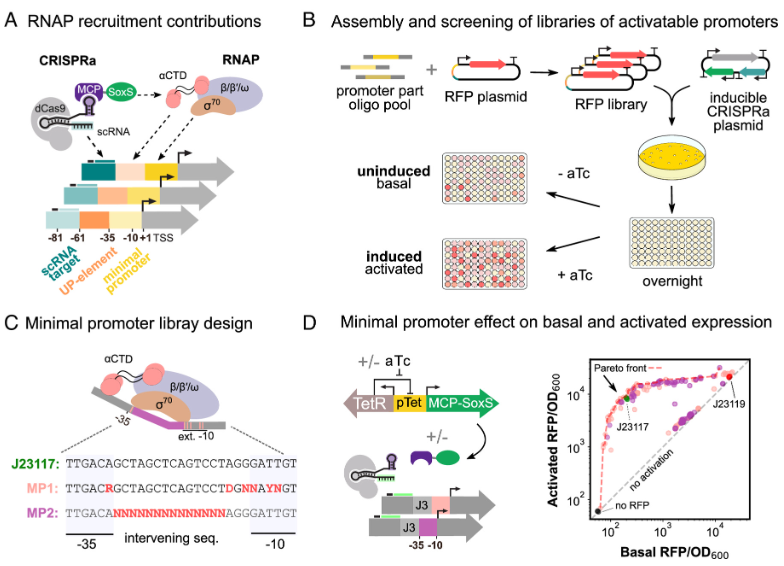
RNAP promoter recognition is enhanced by the AT-rich UP elements located upstream of the minimal promoter, which anchors the α-subunits of RNAP. For effective CRISPRa, RNAP should only be recruited to the promoter in the presence of target sequence-specific scRNA. Therefore, for the transcriptional activation of SoxS, the dynamic range could be improved by reducing the interaction between RNAP-UP elements and reducing the basal expression level. The researchers designed five UP element libraries to contain increasing GC content by mutagenizing the AT-rich E. coli consensus sequence. As expected, consensus UP elements and AT-rich library had the highest basal expression levels. These libraries exhibited only a three-fold activation effect compared to the more GC-rich libraries.
The researchers integrated three previously characterized protein-protein heterodimerization domains into the CRISPRa system. Fusion of these heterodimerization domains to SoxS and MCP enables the conditional recruitment of SoxS to the CRISPRa complex. MCP-SZP6 and SoxS-SZP5 domains were generated for SYNZIP-CRISPRa, MCP-ABI and SoxS-PYL1 domains for ABA-CRISPRa, and MCP-GAI and SoxS-GID1 for GA-CRISPRa. In the original J3 promoter, a 5.7-fold activation was observed when the cognate SYNZIP was fused to the C-terminus of MCP (MCP-SYNZIP) and SoxS (SoxS-SYNZIP). This combination of orientations provides optimal activation. Whereas the activation rate was 2.8-fold when MCP was fused in the opposite direction (SYNZIP-MCP and SoxS-SYNZIP), and 1.4-fold when SoxS was fused in the opposite direction (MCP-SYNZIP and SYNZIP-SoxS). The folding activation of ABA-CRISPRa was also maximized when SoxS was located at the N-terminus. For ABA- and GA-CRISPRa, only C-terminal MCP fusions were tested due to the MCP-SYNZIP results and the precedent of using C-terminal MCP fusions in the CRISPRa system.
The researchers constructed two types of input-responsive pathways to explore the application of the CRISPRa system in the multi-input and multilayer input processing: an AND-like logic gate and a CRISPRa cascade pathway. The scRNA dose-response curve of the CRISPRa system was first characterized. ABA-CRISPRa is highly specific for its target ligand, with no significant activation when GA is present. GA-CRISPRa showed a 3.1-fold cross-activation in the presence of ABA, consistent with reports from eukaryotes. Nevertheless, GA-CRISPRa remained three-fold highly specific for its counterpart ligand, resulting in a 10.5-fold increase in its activation in the presence of GA. These results suggest that the ABA- and GA-CRISPRa systems can be used for orthogonal gene regulation.
Natural biological systems have evolved GRNs containing a variety of activatable promoters, capable of dynamic responses to changing environmental conditions. Activatable promoter engineering enables dynamic regulation of expression levels of large networks of target genes. The CRISPRa/i GRNs can be integrated with existing medical diagnostics to enable complex multi-stimulus signal processing. Going forward, this work could facilitate the development of cellular synthesis and engineered biomaterials, opening up new possibilities for synthetic biology and biotechnology applications.
Reference:
Alba Burbano, D., et al. Engineering activatable promoters for scalable and multi-input CRISPRa/i circuits. PNAS. 2023, 120(30): e2220358120.