Photosynthesis is the basis of life, and in eukaryotes, it occurs in chloroplasts. It is generally believed that chloroplasts originated from endosymbiosis between photosynthetic prokaryotes and heterotrophic eukaryotes that began more than 1 billion years ago. Improving photosynthetic efficiency is considered an important goal of crop improvement. However, in order to design this key process, it is necessary to improve the understanding of the photosynthesis gene regulatory network.
Chloroplast biogenesis depends on the regulation of transcription factors, and GOLDEN2-LIKE (GLK) family transcription factors are its main regulators. But the glk mutant still contains residual chlorophyll, indicating that other proteins are involved. Because forward genetics failed to identify these proteins, Julian M. Hibberd et al. of the University of Cambridge believed that genetic redundancy hindered their identification, and the analysis of species with more compact genomes such as Marchantia polymorpha could circumvent this problem.
On September 5, 2024, Cell published online a research article titled "MYB-related transcription factors control chloroplast biogenesis" by Julian M. Hibberd et al. of Cambridge University, reporting that through the study of the moss M. polymorpha and the angiosperm Arabidopsis thaliana, it was found that MYB-related transcription factors (RR-MYBs) regulate chloroplast biogenesis and photosynthesis gene expression.
In order to identify transcription factors that regulate chloroplast biogenesis, Julian M. Hibberd et al. re-examined, gene edited and phenotypic analyzed the public RNA sequencing data, and identified two RR-MYB-related transcription factors (MpRR-MYB5 and MpRR-MYB2) that regulate chloroplast biogenesis and photosynthesis gene expression.
Functional analysis showed that MpRR-MYB5 and MpRR-MYB2 have a synergistic effect in regulating chloroplast biogenesis. The double mutant Mprr-myb5,2 has very limited chloroplast development, significantly reduced chlorophyll content, smaller chloroplasts, and poorly developed thylakoids. MpRR-MYB5 and MpRR-MYB2 co-regulate chloroplast biogenesis with MpGLK. Chloroplast development in the MpGLK mutant is also affected, but to a lesser extent than in the Mprr- myb5,2 double mutant. In the Mpglk,rr-myb5 double mutant, chloroplasts are smaller, thylakoid membranes are fewer, and grana stacking is reduced. MpRR-MYB5 and MpRR-MYB2 regulate the expression of genes related to chlorophyll biosynthesis and photosynthesis. Their mutations lead to reduced transcript abundance of these genes and reduced expression of photosynthesis-related proteins.
DNA affinity purification and sequencing (DAP-seq) revealed that MpRR-MYB5 and MpRR-MYB2 can bind to the promoters of photosynthesis genes and activate their expression. Studies on homologs in Arabidopsis thaliana (AtMYBS1 and AtMYBS2) verified the conservation of MYB-related transcription factor functions.
The identification of MYB-related transcription factors (RR-MYBs) fills the gap in the understanding of the regulatory mechanism of chloroplast development and provides a new perspective for further studying the formation and function of chloroplasts.
RR-MYBs play a vital role in chloroplast biogenesis, and their targets are broader than the known GLK transcription factors, which provides new potential targets for improving crop photosynthesis efficiency and yield through genetic engineering or breeding, and helps promote crop improvement and agricultural development.
The complex interactions and synergistic regulatory relationships between RR-MYBs and GLK transcription factors will prompt scientists to further study the complexity of gene regulatory networks in plants and how these networks are regulated in response to environmental and developmental signals.
From an evolutionary perspective, RR-MYBs are ubiquitous in terrestrial plants and have conserved functions in different species, which helps to deeply understand how plants adapt to environmental changes and develop complex physiological characteristics during evolution.
Chloroplast biogenesis is closely related to processes such as nitrogen and sulfur assimilation, biosynthesis of amino acids, fatty acids and carotenoids. The study of RR-MYBs may provide clues for the study of these related biological processes and promote a comprehensive understanding of the overall physiological metabolism of plants.
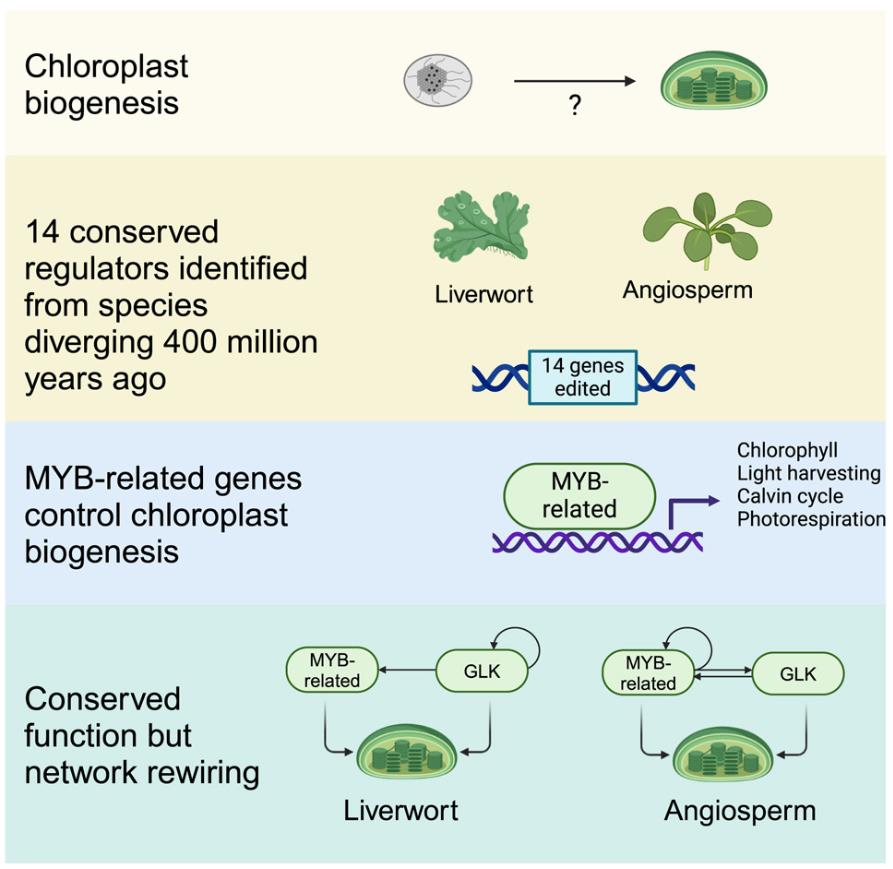
Figure 1. MYB-related transcription factors control chloroplast biogenesis. (Frangedakis, et al., 2025)
Future studies need to further clarify the precise architecture of the gene regulatory network involving RR-MYBs and GLK, including defining the spatial and temporal distribution of their targets through methods such as ChIP-seq, understanding how they are regulated by developmental and environmental signals, and exploring the role of RR-MYBs in their relationship with light signaling networks and hormone networks, as well as interactions with other proteins and post-translational regulation.