Allopolyploid Brassica species Brassica napus and mustard (Brassica juncea) have become important oilseed crops due to increasing global demand for edible oils. The seed meal left after oil extraction is rich in protein and is a promising substitute for soybean-based protein feed for livestock. However, significant amounts of glucosinolates are present in seeds, and reducing seed glucosinolate content (SGC) to canola-quality levels of less than 30 μmoles per gram of dry seed weight (DW) has been a major goal in rapeseed and mustard breeding. Recently, Naveen C. Bisht’s research group from the National Plant Genome Research Institute of India published a paper entitled "Targeted editing of multiple homologues of GTR1 and GTR2 genes provides the ideal low-seed, high-leaf glucosinolate oilseed mustard with uncompromised defence and yield" in Plant Biotechnology Journal. Targeted editing of multiple BjuGTR1 and BjuGTR2 homologues in B. juncea using the CRISPR/Cas9 system improved oil and seed meal quality without compromising the plant protection provided by glucosinolates.
For the study, Varuna, a B. juncea line high in seed glucosinolates, a giant variety belonging to the Indian mustard gene pool, was selected for gene editing. Varuna's GTR1 and GTR2 genes each contain six functional homologues. To achieve a large reduction in SGCs, we aimed to simultaneously target the majority of BjuGTR1 and BjuGTR2 homologues. A scan of BjuGTR homologues in CRISPR-P V2.0 identified a highly conserved 20 nucleotide (nt) region. From this region, 3 guide RNAs (gRNA1, gRNA2, and gRNA3) were designed, which could target a total of 9 BjuGTR homologues with 100% similarity. BjuGTR1-A3 and BjuGTR1-B3 displayed sequence differences in their corresponding Protospacer Adjacent Motif (PAM) sites (AGG to AGA) and were therefore not considered for editing in the study. The target sites of gRNA1, gRNA2, and gRNA3 mapped to the second exon of the respective BjuGTRs, corresponding to the second transmembrane domain in the encoded BjuGTR proteins. Varuna was transformed following the Agrobacterium-mediated genetic transformation protocol. A total of 37 primary transformation events (T0 generation) were generated and the SGC in T1 seeds collected from each T0 event was estimated using HPLC. SGC in GTR1::GTR2(GEd) line T1 seeds ranged from 6.21 to 145.88 μmoles/g DW, while SGC in wild-type (WT) Varuna was 146.09 ± 4.59 μmoles/g DW. Thirty of 37 T0 events showed a significant reduction in SGC, bringing them at or below the Canola quality standard of <30 μmoles/g DW. All major aliphatic glucosinolate fractions (sinigrin, gluconapin, and glucobrassicanapin) were significantly reduced in T1 seeds from GTR1::GTR2(GEd) events compared to WT Varuna.
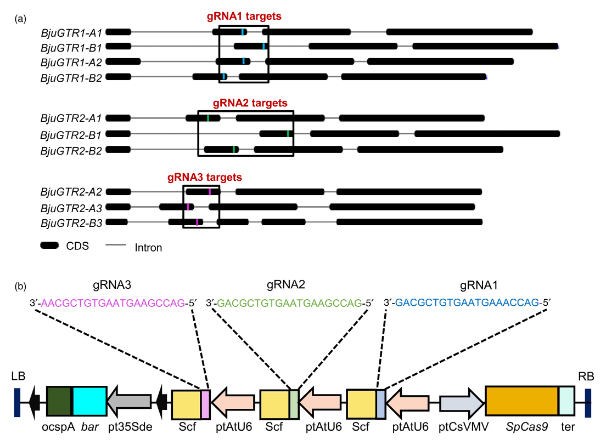
The dramatic reduction of SGCs in GTR1::GTR2(GEd) events prompted investigators to screen for CRISPR/Cas9-induced mutations in target BjuGTR homologues. A total of 22 representative T0 events were selected for screening of mutations in BjuGTR1 and BjuGTR2 homologues. Across all 22 T0 events, the editing frequencies of the 9 BjuGTR homologues ranged from 86% to 100%, showing an overall mutational efficiency of >84%. BjuGTR2-B3 showed the lowest mutation frequency (31.81%), which may be due to its mismatch with the three gRNAs used. Due to the nature of CRISPR/Cas9, all mutations detected were located 3-4nt upstream of the PAM site. Approximately 90% of mutations were found to be single nucleotide insertions, resulting in frame-shift mutations. Biallelic homozygous (BHo) and biallelic heterozygous (BHt) were the most common mutations observed in BjuGTR in T0 events. Mutation profiles of all BjuGTR homologues were compared in T0 events with low SGC and high SGC. Sequence analysis revealed that low SGC lines exhibited frame-shift mutations in most alleles of the target BjuGTR1 and BjuGTR2 homologues, resulting in complete loss of gene function. In T0 events containing high SGCs, most BjuGTR2 homologues were either unedited or in a monoallelic (Mo) edited state.
In conclusion, this work clearly demonstrates that simultaneous editing of multiple (but not all) BjuGTR1 and BjuGTR2 homologues can generate low-SGC, high-LGC B. juncea lines with unaffected defence and yield performance. The benefits of high leaf glucosinolate content were demonstrated by testing the performance of GTR-edited lines against a necrotrophic pathogen (Sclerotinia sclerotiorum) and a common pest (Spodoptera litura). High leaf glucosinolates in the GTR-edited lines helped them perform better or comparable to WT Varuna against the tested pests and pathogens. Also showed a significant advantage over Canola quality lines, which had reduced glucosinolates in all parts of the plant.
Reference:
Mann, A., et al. Targeted editing of multiple homologues of GTR1 and GTR2 genes provides the ideal low-seed, high-leaf glucosinolate oilseed mustard with uncompromised defence and yield. Plant Biotechnol J. 2023 Aug 4. doi: 10.1111/pbi.14121.