Accurate protein subcellular localization detection is necessary for the targeting and functional characterization of the proteins of interest. Lifeasible provides two powerful approaches for precise tracking of protein residing in plant cells: the technically established immuno-labeling approach, and the advanced live-cell imaging approach. Our expertise on both approaches ensures subcellular tracking of a large variety of proteins, in differential plant tissues, across species.
Immuno-labeling
The Immuno-labeling technique enables precise detection of protein localization by utilizing antibodies that specifically bind to the proteins of interest (antigen). Commonly, the immune-labeling process employs two types of antibodies: a biochemically engineered primary antibody that can specifically bind to the antigen, as well as a tagged secondary antibody that can covalently bind to the primary antibody (1). The tag that harbored by the secondary antibody allows specific detection and/or visualization of the proteins of interest (Figure 1). Adapting to differential detecting techniques, the tag can be a fluorescent reporter, such as green fluorescent protein (GFP), an enzyme that can produce a signal compound, or electron-dense particles such as gold beads (2).
With years of experience in antibody developing and engineering, Lifeasible proudly provides advanced services for the biotechnological engineering and processing of antibodies against plant proteins. Moreover, with the support of the extensive expertise of our collaborators, we provide a variety of cutting-edge techniques for the detection of immuno-labeled proteins. Adapting to different plant species, specific tissues and cell types, detecting of targeted proteins can be realized via optic microscopy, fluorescence microscopy, electron microscopy or atomic force microscopy.
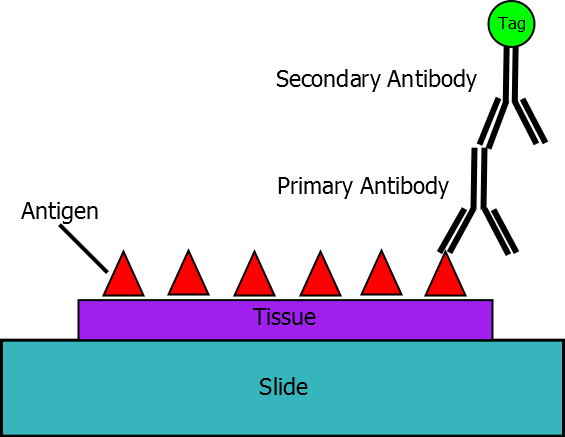 Figure 1. A schematic cartoon of immuno-labeling (from Wikipedia).
Figure 1. A schematic cartoon of immuno-labeling (from Wikipedia).
Live-cell imaging
Comparing to immune-labeling, which requires fixation of plant tissue samples, live-cell imaging allows visualization of targeted proteins in living cells in their native states (3). Therefore, this technique has become increasingly popular, as it provides better understanding of the biological functions of plant proteins. Specifically, the advantages of live-cell imaging are:
The live-cell imaging process mainly includes three steps:
Reference