Coprinopsis cinerea taxonomically belongs to Basidiomycotina, Hymenomycetes, Agaricales, Psathyrellaceae, Coprinus. In 2003, the whole genome of Coprinopsis cinerea was sequenced and its haploid nuclear genome is about 37.5 Mb with 13 chromosomes and is expected to contain 13,000-15,000 genes. Although the edible value of Coprinopsis cinerea is limited, as a good model organism to study the growth mechanism of stretcher fungi, it can help researchers understand the growth, development, and reproduction mechanism of other edible and medicinal fungi that are difficult to cultivate in the laboratory, and guide the production of edible fungi and improve the yield of edible fungi.
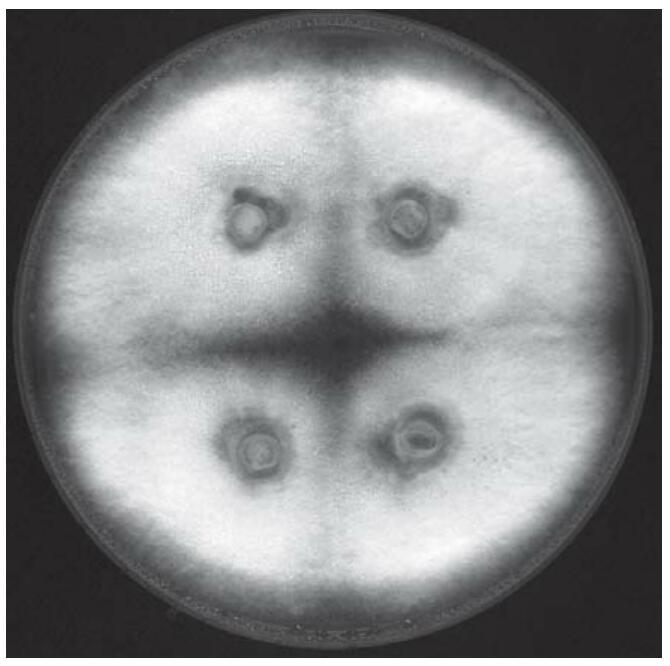 Figure 1. YMG/T agar plate inoculated with four mycelial agar pieces of C. cinerea FA2222 after six days of growth at 37°C in the dark. (Dörnte B, et al. 2012)
Figure 1. YMG/T agar plate inoculated with four mycelial agar pieces of C. cinerea FA2222 after six days of growth at 37°C in the dark. (Dörnte B, et al. 2012)
In the genetic transformation of Coprinopsis cinerea, exogenous genes are integrated into the genome of Coprinopsis cinerea by random insertion. The integration of exogenous gene fragments into the host genome is dependent on the DNA double-strand break repair mechanism of the host cell. Appropriate selection of markers is a prerequisite for genetic transformation and can effectively reduce false positives and accelerate the progress of the project. Nutrient-deficient screening involves transferring nutrient marker genes into the genome of a nutrient-deficient recipient strain to make it exhibit the phenotype of a wild-type strain.
Lifeasible has nutrient-deficient markers such as trp-1 for tryptophan synthase, argB for ornithine carbamoyltransferase, trpC for Penicillium chrysogenum and Aspergillus niger, and pyrG for Aspergillus oryzae and Penicillium chrysogenum.
Lifeasible can use nutrient-deficient markers to isolate specific nutrient-deficient receptor strains. We provides Coprinopsis cinerea tryptophan synthase-deficient strains, pab1 nutrient-deficient strains, etc. that can be used as receptor strains, and has pCC1001, pTPHT-6, pCRG6HT-1, pCRG7HT-1, and pPABl-2 plasmids to construct PABA nutrient-deficient plasmids were used as screening markers to construct the genetic transformation system of Coprinopsis cinerea. Besides, we also constructed Hygromycin screening plasmids to construct the genetic transformation system of Coprinopsis cinerea by the genetic transformation method of Agrobacterium-mediated transformation with Hygromycin (HygR) as the screening marker.
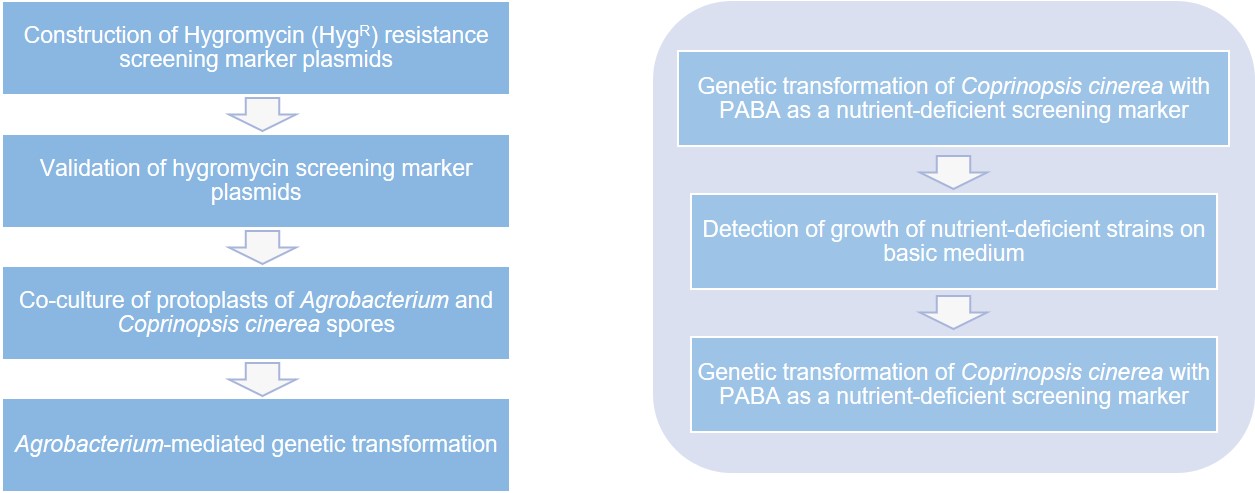
We not only construct the genetic transformation system of Coprinopsis cinerea, but we also provide the following services.
References: