Fish genome research uses large-scale sequencing to obtain genome sequence information about species. It then elucidates the relationship between fish biological properties, phenotypic variation, and genome structure, which has excellent potential for application of fish germplasm identification, genetic breeding, animal nutrition, environmental toxicology, and disease control. With the development of whole genome sequencing technology, ATAC-seq technology has become a powerful tool for fish epigenetic studies.
ATAC-seq technology identifies and cleaves open chromatin regions by transposases, thereby obtaining the regulatory sequences of all active transcripts in the genome. ATAC-Seq can help you analyze how chromatin conformation and other factors affect gene expression and is widely used in popular research areas such as fish embryo development, sex reversal, fin regeneration, disease mechanisms, and genomic selection for breeding.
Lifeasible has been deeply involved in the sequencing field for many years and has a unique profile for animal sequencing. We have built a complete epigenome sequencing platform to provide you with an innovative, flexible, and scalable fish ATAC-seq solution to help customers infer the accessibility of fish chromatin regions and calculate the regional location of transcription factor binding sites and nucleosomes.

Chromatin structure plays a key role in regulating gene expression precisely. We used ATAC-seq to examine open chromatin regions during sex reversal in orange-spotted grouper (Epinephelus coioides). We combined it with RNA-seq to analyze the binding sites of open chromatin regions to transcription factors (TFs). Several sex-specific or cell-specific marker genes that may trigger sex reversal was identified, supporting the study of the regulatory mechanisms of sex reversal in fish.
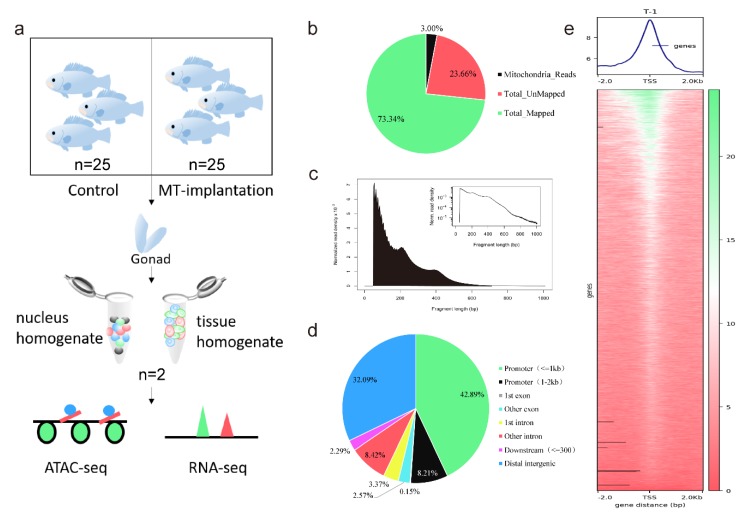 Figure 1. Overview of ATAC-seq detection of orange-spotted groupers. (Wu, X, et al. 2020)
Figure 1. Overview of ATAC-seq detection of orange-spotted groupers. (Wu, X, et al. 2020)
We performed epigenome sequencing of purified fibroblasts from uninjured and regenerated caudal fins by ATAC-seq technology. In combination with RNA-seq, identifying novel noncoding regulatory sequences near induced and/or essential genes during fin regeneration verified distinct domains of directed expression for each confirmed tissue regeneration enhancer element (TREE).
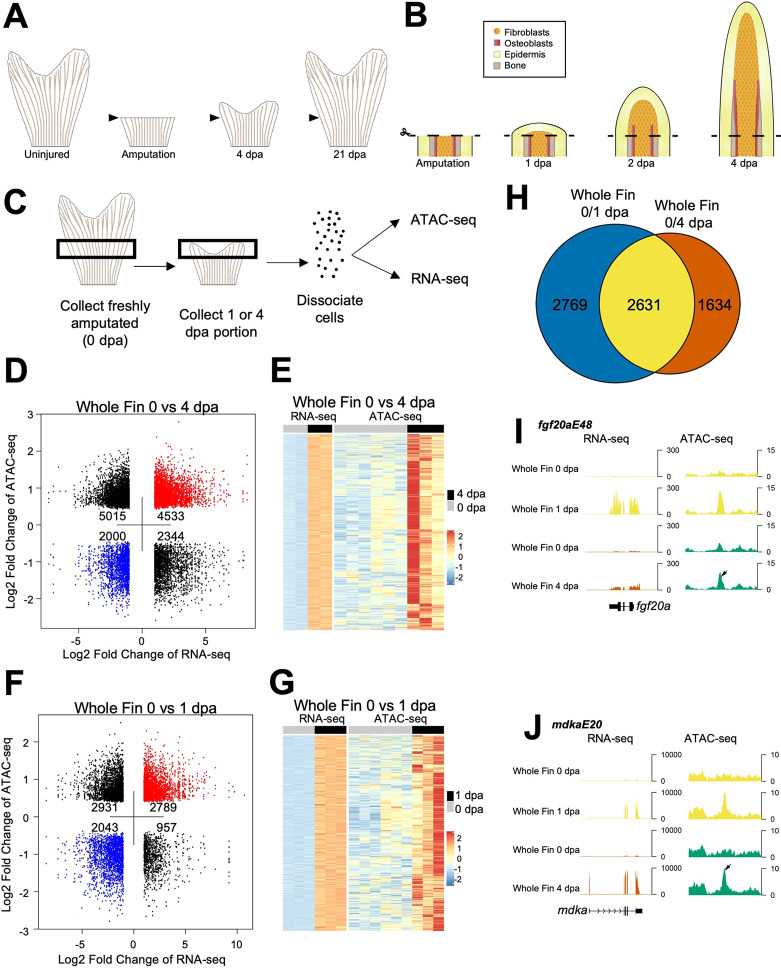 Figure 2. Chromatin accessibility profiling of regenerating zebrafish fins. (Thompson, J. D, et al., 2020)
Figure 2. Chromatin accessibility profiling of regenerating zebrafish fins. (Thompson, J. D, et al., 2020)
Orofacial cleft (OFC) may include cleft lip, palate, or both. To analyze human OFC risk-associated single nucleotide polymorphisms (SNPs), we analyzed zebrafish periderm enhancer candidates by ATAC-seq technique using zebrafish as a model. The results showed that 14 OFC risk-associated SNPs were located in the peridermal enhancers that regulate KRT18 / KRT8 expression.
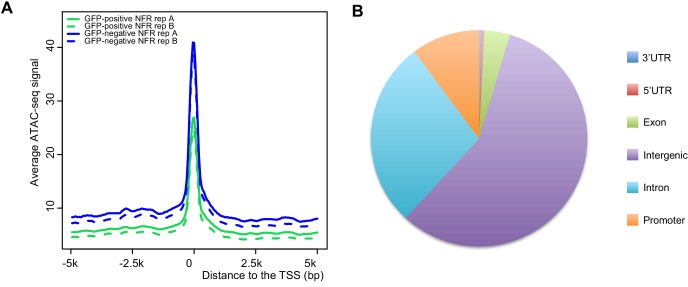 Figure 3. Annotation of the ATAC-seq peak relative to the transcription start site. (Liu, H, et al. 2020)
Figure 3. Annotation of the ATAC-seq peak relative to the transcription start site. (Liu, H, et al. 2020)
Please note: Ensure sufficient dry ice when transporting samples to avoid temperature rise to destroy cell activity.
Lifeasible's animal-oriented sequencing technology platform can provide you with one-stop fish ATAC-seq services. Our scientific protocol design, strict quality control management, and professional analysis team can ensure that every step of the process will be completed with excellence. For questions, inquiries, or collaboration, please feel free to contact us.
References