Biotic stress is caused by many factors, including infection by fungi, bacteria, viruses, and nematodes, and competition with weeds. Biotic stress can lead to severe losses in forage yield and quality, making control of biotic stresses significant. Using fungicides and insecticides in forage grasses is limited due to their cost and human and environmental safety considerations. One of the most effective strategies to control pests and diseases is to breed resistant forage varieties. A promising grass variety must contain genes for resistance to biotic stresses. Forage grasses have many strategies to protect themselves from biotic stresses, such as allergic reactions and the production of plant antitoxins. The isolation of genes for biotic stress resistance in forage grasses and the analysis of molecular mechanisms of biotic stress resistance have become a focus of interest.
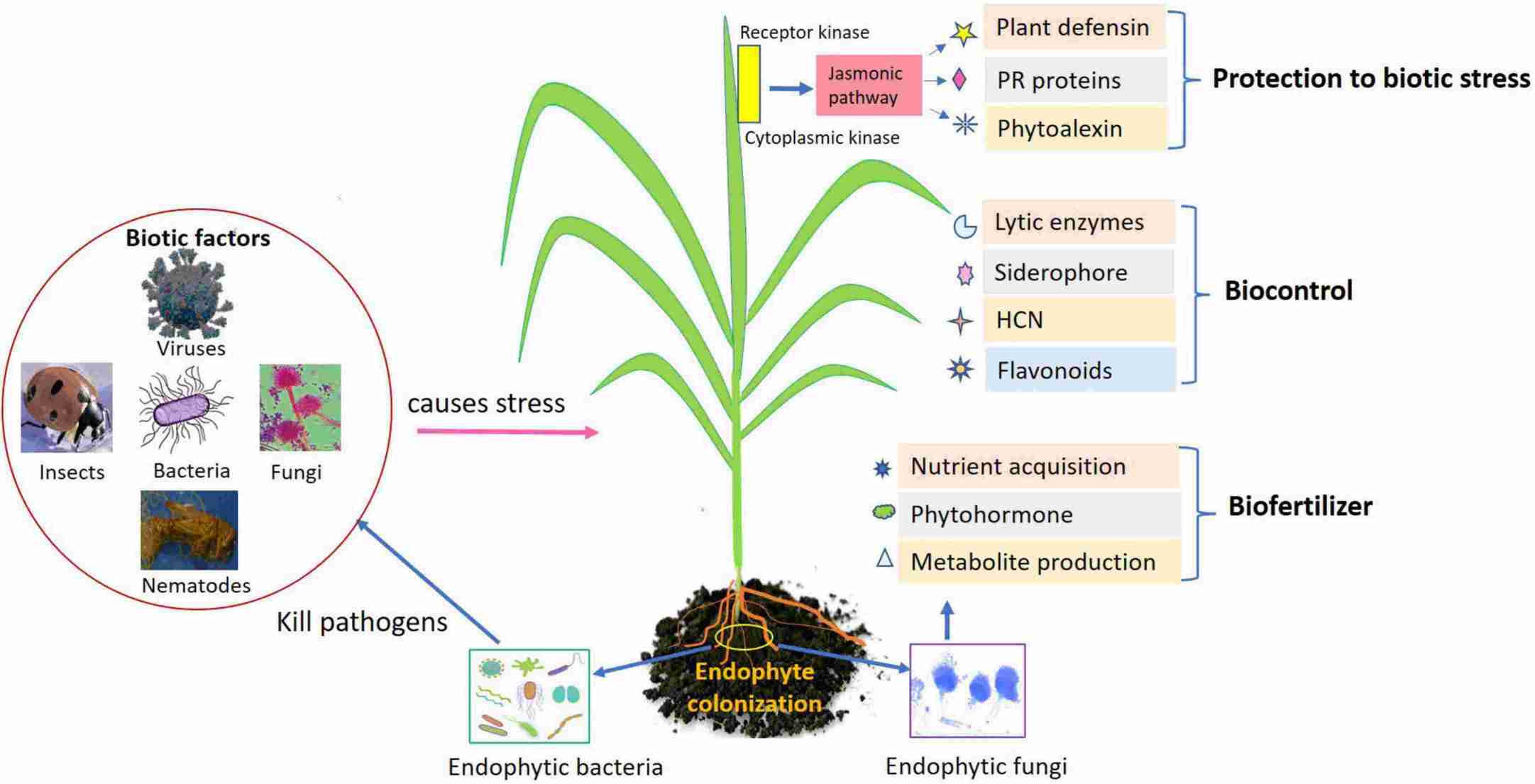 Fig.1. Role of endophytes as biofertilizers, biocontrol, and biotic stress management in agricultural crops. (Chaudhary P et al., 2022)
Fig.1. Role of endophytes as biofertilizers, biocontrol, and biotic stress management in agricultural crops. (Chaudhary P et al., 2022)
Forage grasses frequently encounter many potential pathogens that can infect plants and cause diseases. Our team of experts has done a lot of work in this field. As an ideal partner for forage research, Lifeasible offers genomic analysis services for disease resistance in forage by identifying genes that confer tolerance to biotic stresses in forage. We isolated and characterized more than 30 disease-resistance genes in forage grasses and explored novel resistance genes. In addition, we offer forage resistance loci studies focusing on identifying resistance genes against bacteria, viruses, fungi, nematodes, and insects.
We aim to improve forage varieties by identifying genes for resistance to biotic stresses and to help our customers breed forage varieties resistant to biotic stresses. Please see below for our custom genomic analysis solutions for disease resistance in forage grasses.
Resistance to crown rust is a very important trait for forage grasses because the broad host range of crown rust includes the genera Lolium, Festuca, Agropyron, Agrostis, Paspalum, Phleum, Poa, and Puccinellia. We used amplified fragment length polymorphisms for bulk segregation analysis of resistance to crown rust genes.
We pyramid the resistance genes of the target forage species by identifying several master or minor genes. We provide DNA markers closely linked to known resistance loci for linkage analysis to explore new resistance loci. It is important to note that to determine the specificity of the resistance genes, lines carrying a single resistance gene in the pure state are required as test lines for seed identification.
We can introduce resistance genes from wild species to develop novel resistance varieties in forage grasses.
We can easily use DNA markers closely linked to resistance loci to develop forages with the target gene. This technique of ours helps plant breeders to be able to monitor the frequency of resistance genes and the genetic diversity of resistance loci in the breeding population. In addition, we provide SSR markers to analyze the high diversity of resistance loci to help stabilize forage grass resistance to various pathogens.
We provide map-based cloning and transposon-based gene marker technologies to isolate resistance genes from forage grasses. In addition, we develop cutting-edge genomic covariance methods for large regions to achieve efficient isolation of resistance genes in forage grasses.
We provide microarray technology to simultaneously analyze the expression of thousands of disease-resistance genes in forage grasses. We construct rich cDNA libraries to obtain a comprehensive set of genes associated with disease resistance.
We provide forage symbiotic genomic services to identify genes involved in forage colonization and the nutrient supply of endophytic fungi. Our genomic tools and knowledge will support the development of technologies to manipulate linkages between grass endophytes, improve forage tolerance to organisms, and alter the host specificity of grass endophytes.
Resistance to biotic stresses is a fundamental characteristic of forage grasses. We focus on discovering and validating candidate genes for disease resistance in forage grasses. Working closely with international breeders, we use advanced forage technologies to help our customers breed resistant and durable varieties to biotic stress. For more information or to discuss in detail, please contact us.
Reference