Insects are the most diverse and ancient group of living organisms. Insects are relevant to humans, as pests that cause great production and health hazards and as taxa that provide key service functions to agroecosystems.
With the rapid development of sequencing and analysis technologies, entomological research has also moved into the genomic era. Entomologists use various omics research tools, such as genome, transcriptome, and proteome, to solve problems in entomological research from the perspective of systems biology. Epigenetics, which studies chromatin conformation and gene expression, has received much attention from scientists in recent years. Therefore, ATAC-seq is widely used as a mainstream tool for chromatin conformation studies in insects.
Lifeasible has been in insect sequencing for many years and provides entomologists with innovative, flexible, and customizable insect ATAC-seq solutions. Our epigenome sequencing solutions bring new perspectives to entomological research, helping our customers gain insight into the genes that regulate insect growth, development, metamorphosis, rhythm, evolution, and reproduction.

Taking butterflies as the research object, we randomly inserted and cleaved the exposed chromatin DNA in the nucleus using the super-active Tn5 transposase and then ligated it with specific sequencing adaptors. DNA fragments linked to adaptors were isolated for high-throughput sequencing and analysis, yielding information on genome-wide open chromatin regions at specific developmental stages and potentially active transcription factors and their target genes.
To gain a comprehensive understanding of the open chromatin regions of the bumblebee genome and to detect regulatory elements at the genome-wide level, we performed ATAC-seq on Bombus terrestris samples at four developmental stages: egg, larva, pupa, and adult. 36,390 chromatin-accessible regions, including shared and specific stages, were finally identified, contributing to the understanding of bumblebees. The chromatin conformation throughout development was determined.
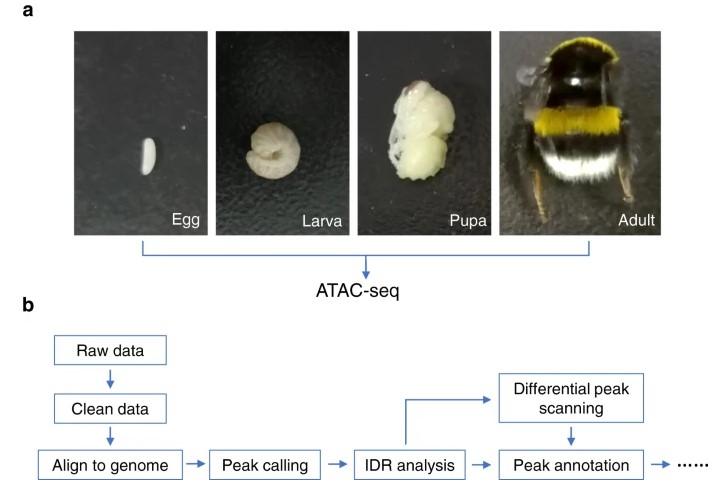 Figure 1. Flow chart for identification of bumblebee chromatin accessibility by ATAC-seq. (Zhao, X, et al. 2020)
Figure 1. Flow chart for identification of bumblebee chromatin accessibility by ATAC-seq. (Zhao, X, et al. 2020)
Development of the Drosophila embryo is controlled by gene regulatory networks (GRNs) that unfold along the anterior-posterior (AP) and dorsoventral (DV) axes. Their different concentrations along the body axis provide location-specific information that translates into different transcriptional outputs in each cell. We determined chromatin accessibility at spatial and temporal resolution using ATAC-seq. The results suggest a positive interaction between regulatory mechanisms and the local chromatin environment of the axis patterning enhancers.
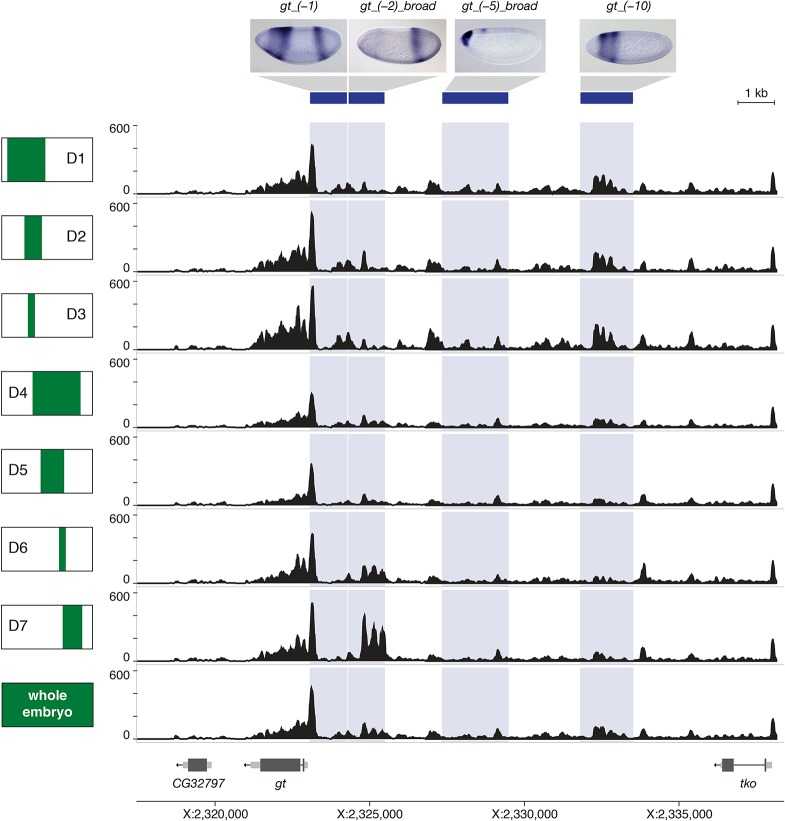 Figure 2. Regional differences in chromatin accessibility. (Bozek, M, et al. 2019)
Figure 2. Regional differences in chromatin accessibility. (Bozek, M, et al. 2019)
Please note: Ensure sufficient dry ice when transporting samples to avoid temperature rise to destroy cell activity.
Lifeasible's animal-oriented sequencing technology platform can provide insect ATAC-seq services to meet our client's needs. Our extensive project experience, scientific protocol design, and strict quality control management ensure that every step of the process is completed with excellence. Please feel free to contact us for questions, inquiries, or collaboration.
References