Pigs have unique biological characteristics and occupy an essential position in livestock production and medical research. The whole genetic series mapping of pigs has been completed in recent years, and the pig gene sequence and chromosome structure are highly isomorphic with humans. The biological traits, reproduction, development, and other life histories of pigs are highly comparable to those of humans. Moreover, the disease types, pathological processes, and mechanisms of pigs are like those of humans. Therefore, pigs are widely used as large model animals.
More importantly, swine disease models are highly controllable, can simulate multifactorial pathology, are highly operable in vivo and in vitro, and can monitor and observe treatment effects in vivo and in real-time, etc. The proportion of biomedical and biotechnology research using pigs as models has been rising in recent years. Therefore, the application of ChIP-seq technology on pigs has received more and more attention from scientists.
Lifeasible, a highly regarded biotechnology company, has established a dedicated ChIP-seq platform for animals to provide professional pig ChIP-seq services to facilitate research related to pig epigenetics.

Our pig ChIP-seq technology service is simple and fast, with high volume and quality of chromatin obtained and high quality of peak maps from ChIP-seq experiments. The final data analysis can predict the function of specific proteins, histone chromosome binding profiles, and epistatic modifications of DNA, which can deepen studies on disease pathology, drug screening, etc.
We sequenced cis-regulatory elements from different tissues of four breeds of pigs using ChIP-seq technology, made a cis-regulatory element map of pigs, improved the understanding of functional elements in the pig genome, and found that the conservativeness of cis-regulatory elements was high between human and pig genomes, even higher than between human and mouse.
lncRNAs can be epigenetically regulated through histone modifications. We identified the genome-wide profiles of H3K4me3 and H3K27ac in the porcine placenta during gestation (the establishment stage of the placental fold) by using the ChIP-seq method. Differentially expressed lncRNAs were analyzed, and several H3K4me3/H3K27ac-lncRNAs affecting porcine placenta development were inferred.
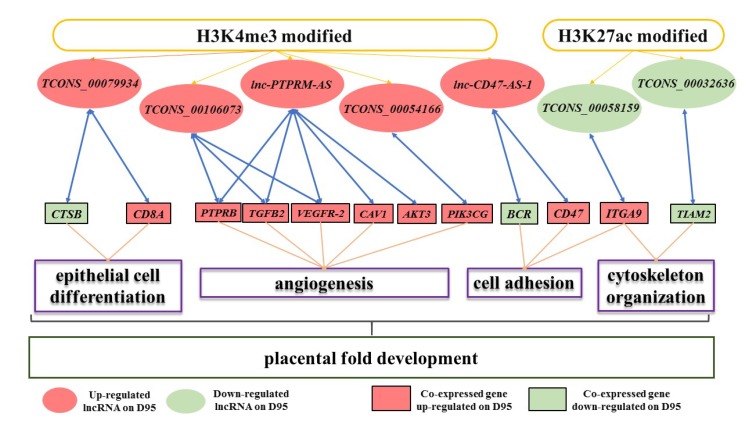 Figure 1. Schematic representation of the potential role of the H3K4me3/H3K27ac-lncRNA-gene pathway in porcine placental development obtained by ChIP-seq. (Deng, D, et al. 2020)
Figure 1. Schematic representation of the potential role of the H3K4me3/H3K27ac-lncRNA-gene pathway in porcine placental development obtained by ChIP-seq. (Deng, D, et al. 2020)
A comprehensive exploration of regulatory elements in the Bama Xiang and Large White Pigs was performed using a ChIP-seq approach targeting H3K4me3 and H3K27ac. ChIP-seq data were analyzed to expand the database of regulatory elements in the H3K4me3 and H3K27ac regions of the porcine pituitary and to compare differences between breeds.
It is recommended to provide two sample preparations, if possible, to ensure the quality and continuity of the experiment.
Lifeasible's animal-oriented sequencing technology platform can provide satisfactory pig ChIP-seq services. Our scientifically designed protocols and strict quality control ensure that every step of the process is completed with excellence, enabling you to conduct scientific research in porcine epigenetics. Please feel free to contact us for questions, inquiries, or collaboration.
References