Animal models are an indispensable tool in biomedical research. The rat is the second most used animal model in the literature. Compared to mice, rats are larger, more aggressive, and more resistant to various diseases. As a commonly used mammalian model organism, the rat has contributed greatly to our study of the function of individual genes, the mechanisms of various diseases, and the efficacy and toxicity of many drugs and chemicals.
The genomes of rat model animals have now been sequenced, and many genes have been found to be conserved between rat model animals and humans. The rat is often used for research in immunology, oncology, physiology, pathology and neuroscience, neurobiology, etc. With the advancement of research and sequencing technology, ChIP-seq is increasingly used in rats.
Lifeasible, as a world-renowned animal and plant biotechnology company, has a professional sequencing team and has built a high-throughput sequencing technology platform which can provide you with a one-stop rat ChIP-seq service. We aim to solve your worries and help you focus on exploring the network mechanism of target protein-DNA interactions.

Lifeasible's rat ChIP-seq technology uses the advanced Illumina sequencing platform to enable fast, efficient reads of high-quality sequencing data to help customers conduct purpose-specific research.
We investigated the relationship between DNA damage, unrepaired DNA accumulation, and neurodegenerative diseases by inducing exogenous DNA damage in rats through X-ray irradiation. ChIP-seq analysis of damaged rat cortical neurons revealed genomic regions rich in γH2AX signaling and close to genes essential for neurodegenerative diseases such as epm2a, serpini1, and il1rpl1.
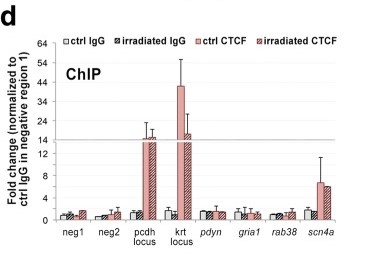 Figure 1. ChIP-qPCR analysis of CTCF enrichment around several γH2AX binding sites defined by ChIP-seq. (Mata-Garrido, J, et al. 2018)
Figure 1. ChIP-qPCR analysis of CTCF enrichment around several γH2AX binding sites defined by ChIP-seq. (Mata-Garrido, J, et al. 2018)
The appearance of inflammatory signals is closely associated with pulmonary arterial hypertension disease. We performed ChIP-seq analysis in a rat pulmonary arterial hypertension (PAH) model and identified various inflammation-related genes, including Tlr5, Ptx3, Ackr3, and Edn1, which can effectively reveal the physiological and biochemical processes associated with inflammation.
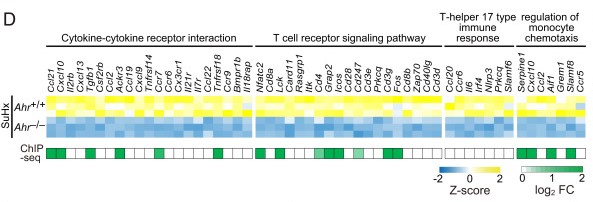 Figure 2. Log2 fold change of enriched genes of ChIP-seq data about cytokine–cytokine receptor interaction. (Masaki, T, et al. 2021)
Figure 2. Log2 fold change of enriched genes of ChIP-seq data about cytokine–cytokine receptor interaction. (Masaki, T, et al. 2021)
Astrocytes are involved in brain learning memory and are associated with neuropsychiatric disorders. We help our clients to test the epigenetic effects of drugs, stimuli, or genetic engineering on cells by performing ChIP-seq analysis on rat astrocytes.
It is recommended to provide two sample preparations, if possible, to ensure the quality and continuity of the experiment.
Lifeasible can perform a wide variety of ChIP-seq analyses in rat cells, and we guarantee to provide researchers with precise services to meet your research needs in gene difference analysis, gene function annotation, and gene protein interaction. We can provide pre-experimental consulting services to provide you with a personalized service plan, so please feel free to contact us if you need further information.
References