Autophagy of Endoplasmic Reticulum Quality Control for ER Stress in Plants
Autophagy functions as a degradation process in the recycling of cellular cytoplasmic contents and the removal of damaged proteins or organelles under unfavorable growth conditions. Since many proteins in endoplasmic reticulum (ER) are membrane proteins, the bulk of multiple protein molecules gathered together cannot be reversed and transported to the cytoplasm for degradation by the proteasome. Therefore, these proteins are generally degraded by the autophagy pathway that can regulate protein stability.
Lifeasible is a leading provider of comprehensive, high-quality services for plant science. Our platform is equipped with state-of-the-art facilities and highly experienced staff in order to assist in areas of autophagy of endoplasmic reticulum quality control. We guarantee credible results for our customers all over the world.
IRE1 in Autophagy of Endoplasmic Reticulum Quality Control
- Autophagy is induced by ER stress in plant cells, IRE1 is the link between these two processes and the kinase function of IRE1, but not its ribonuclease function is required for the induction of autophagy.
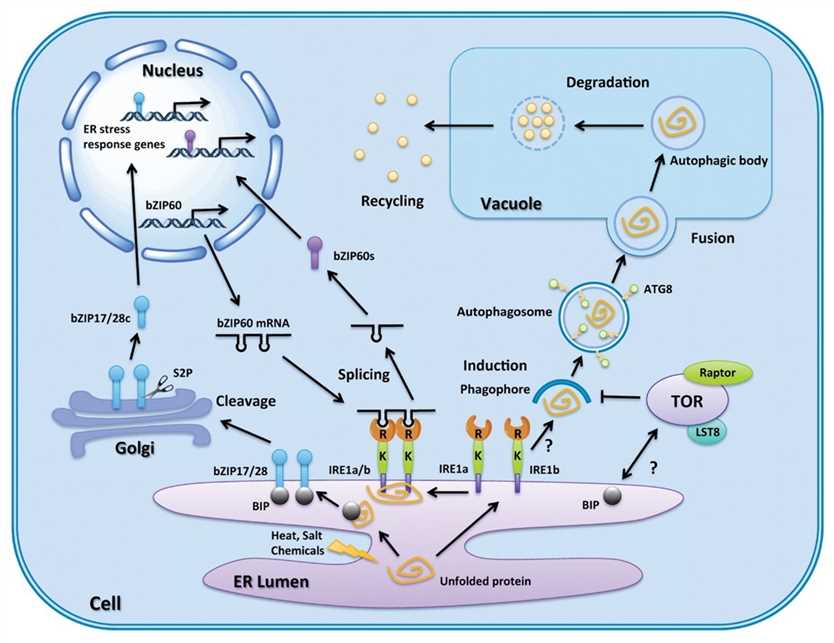 Fig.1. Autophagy and ER stress regulation pathways in plants. (Pu Y and Bassham DC., 2013)
Fig.1. Autophagy and ER stress regulation pathways in plants. (Pu Y and Bassham DC., 2013)
- Lifeasible provides services covering this biological process, including MDC staining and microscopy, the transient transformation of leaf protoplasts, generation of IRE1-FLAG construct and complementation of transgenic plants, electron microscopy analysis, and so on.
- To investigate whether ER stress-induced autophagy is activated via the IRE1, we perform MDC staining in ire1 null mutants compared to wild-type plants to examine the induction of autophagy. Likewise, we analyze the number of autophagosomes per root section of ire1 mutants.
- In addition, we transform leaf protoplasts from wild-type and ire1 plants with GFP-ATG fusion constructs to visualize the induction of autophagy under ER stress by confocal microscopy.
BiP in Autophagy of Endoplasmic Reticulum Quality Control
- A key pathway for autophagy regulation is the TOR (target of Rapamycin) signaling pathway, a negative regulator of autophagy in plants. The TOR pathway stimulates cell growth by promoting anabolic processes and inhibiting catabolic processes, such as biosynthesis and autophagy.
- Lifeasible helps our customers study this process through the following ideas. To demonstrate that BiP may be modified by Thr phosphorylation in response to rapamycin-mediated TOR signaling, we analyze BiP levels by Western blot. To further characterize the modification of BiP, we also attempt to identify the phosphorylation sites sensitive to rapamycin by MS.
Lifeasible provides fast turnaround, high-quality services at competitive prices for customers worldwide. Our advanced technical platforms can help our clients solve the problems they may encounter in research based on years of experience. If you are interested in our services or have any questions, please feel free to contact us or make an online inquiry.
Reference
- Pu Y and Bassham DC. (2013). "Links between ER stress and autophagy in plants." Plant Signal Behav. 8 (6), 24297.
For research or industrial raw materials, not for personal medical use!
 Fig.1. Autophagy and ER stress regulation pathways in plants. (Pu Y and Bassham DC., 2013)
Fig.1. Autophagy and ER stress regulation pathways in plants. (Pu Y and Bassham DC., 2013)