Precisely regulating the expression of endogenous genes through CRISPR/Cas9 gene editing technology is crucial to achieve precise design and breeding of crops. The most widely used application of CRISPR/Cas9 technology in breeding is to knock out target genes, thereby achieving loss-of-function. However, the improvement of many agronomic traits requires gain-of-function alleles, which upregulate the expression of target genes, that is, gene knock-up. Therefore, the development of new methods to upregulate gene expression through gene knock-out can expand existing gene expression regulation tools without changing the gene coding sequence and introducing new genome segments, and provide strong technical support for crop genetic improvement and de novo domestication of wild plants.
On November 9, 2023, Hong Yu's team at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences published a research paper titled "Genome editing of 3′ UTR-embedded inhibitory region enables generation of gene knock-up alleles in plants" online in Plant Communications. This study developed a new method to upregulate the protein expression of genes by knocking out the negative regulatory region in the 3' untranslated region (3'UTR) of plant genes, modified three genes in rice and Arabidopsis, and obtained plants with successfully improved traits.
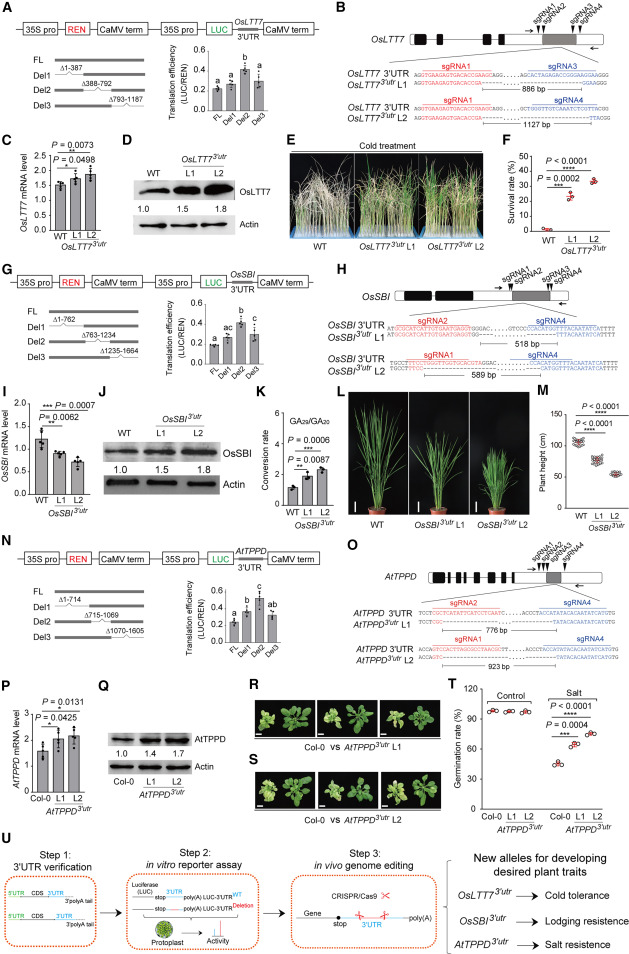
The 3'UTR of eukaryotic genes mainly affects the precise expression of genes by regulating the translation of mRNA. The 3'UTR negative regulatory region usually contains cis-elements specifically recognized by miRNA or forms a relatively stable RNA secondary structure to inhibit translation. Therefore, this study speculates that knocking out the 3'UTR negative regulatory region through CRISPR/Cas9 technology can upregulate the expression of target proteins, thereby creating new gene knock-up alleles to improve plant traits. To test this idea, the authors selected the low-temperature tolerance gene OsLTT7 and dwarfing gene OsSBI in rice, as well as the salt-tolerance gene AtTPPD in Arabidopsis for study. First, the 3'UTR sequence of the target gene was cloned and identified using 3'RACE technology, which is rapid amplification of cDNA ends. Then, a protoplast-based dual-luciferase reporter system was used to screen the regulatory regions that exert the most significant inhibitory effect on the luciferase reporter gene. Finally, four CRISPR/Cas9 sgRNAs were designed and constructed for the 3'UTR target region of each target gene for editing. The authors studied the obtained homozygous mutants without transgenes and found that deletion of the 3'UTR negative regulatory region increased the abundance of target proteins in OsLTT73'utr, OsSBI3'utr and AtTPPD3'utr mutants. OsLTT73'utr, OsSBI3'utr and AtTPPD3'utr plants acquired the traits of low temperature tolerance, dwarfing and salt resistance respectively. Based on this, this study proposes a technical roadmap for creating gene knock-up alleles.
Reference:
Wang, H., et al. Genome editing of 3' UTR-embedded inhibitory region enables generation of gene knock-up alleles in plants. Plant Commun. 2023, Nov 9: 100745.